Help
Submission
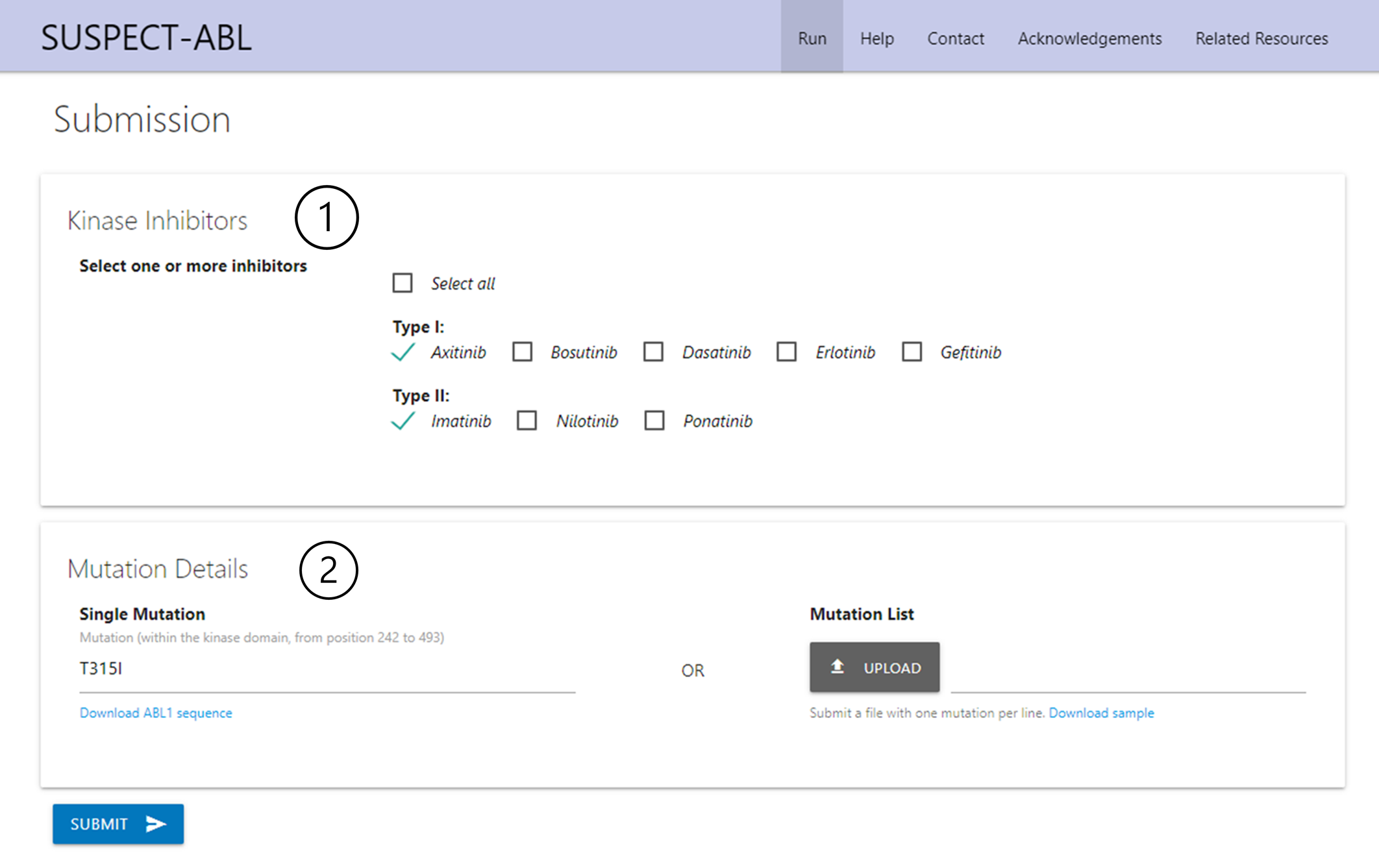
1. Choose one or more inhibitors of interest.
2. Submit mutation: either as a single point mutation format or as a mutation list. FASTA sequence for ABL1 is available for download. Sample lists are available for download showing formatting required.
Results - Single Prediction
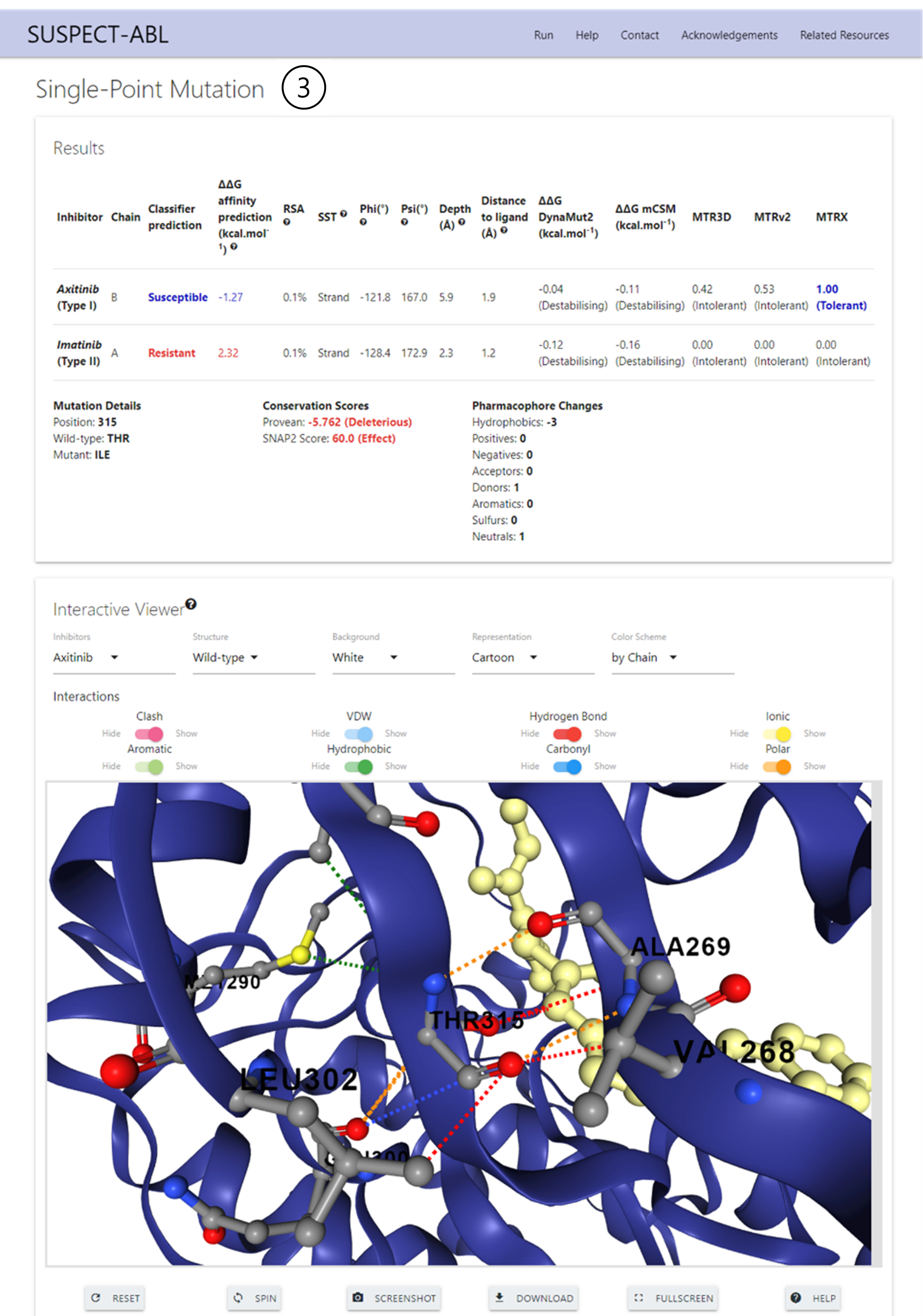
3. Single Point Mutation visualisation:
In the ‘Results’ section, the predictions for the regressor and the classifier are displayed in a table, along with the information about the wild-type environment and several other predicted parameters. Mutation details, conservation score and pharmacophore changes are also shown below the table.
In the 'interaction Viewer' section, users can visualise the non-covalent interactions around the mutation site, and are allowed to select different inhibitors, compare the wild-type and mutant, as well as customize the representation. This viewer also enables molecule rotation, transposition, spinning and zoom, with added screenshot and PyMOL session downloads.
Results - List Prediction
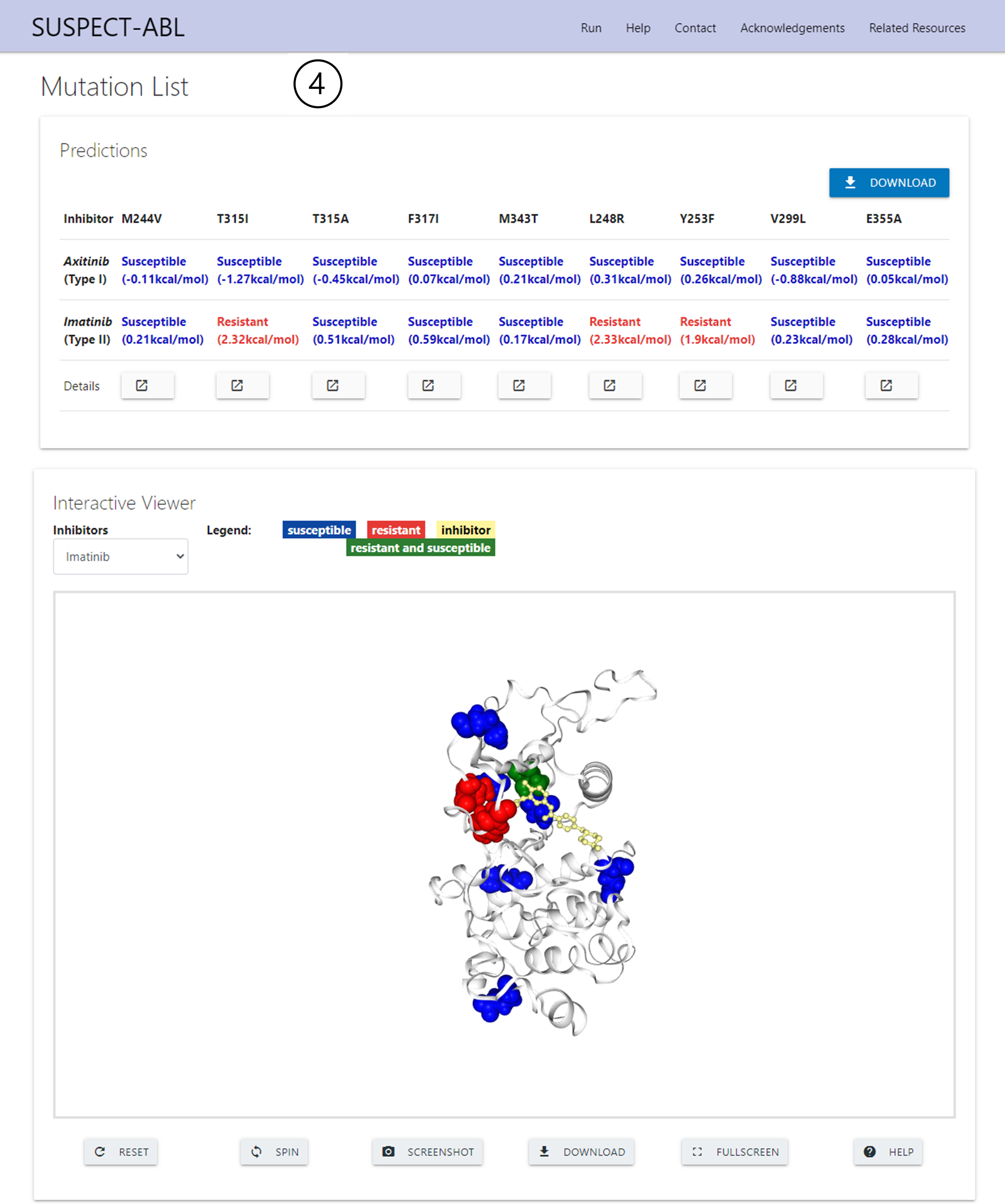
4. Mutation List visualisation:
In the ‘Predictions’ section, the predictions for each drug against each mutation in the input file are shown in the table.
The ‘Details’ row permits the user to access the full details for each mutation submitted, in a similar format to point 3.
The ‘Interactive Viewer’ section visualises how all the mutations within the submitted list map across the ABL1, where resistant mutations are shown in red, susceptible in blue, and any positions submitted having different phenotypes in green. This viewer also enables molecule rotation, transposition, spinning and zoom, with added screenshot and PyMOL session downloads.