SUSPECT-ABL StrUctural Susceptibility PrEdiCTion on ABL
Yunzhuo Zhou, Stephanie Portelli, Megan Pat, Thanh Binh Nguyen, Carlos H.M. Rodrigues, Douglas E.V. Pires, David B. Ascher
Abstract: Kinases play crucial roles in cellular signalling and biological processes. Dysregulation of their activities is associated with diseases, including cancers. The introduction of kinase inhibitors, most notably those targeting ABeLson 1 (ABL1) kinase in chronic myeloid leukemia, has had a significant impact on cancer survival, yet the emergence of resistance mutations can reduce their effectiveness and lead to therapeutic failure. Limited effort, however, has been devoted to developing tools that are capable of accurately identifying potential ABL1 resistance mutations, as well as providing relevant insights into the molecular mechanisms they bestow.
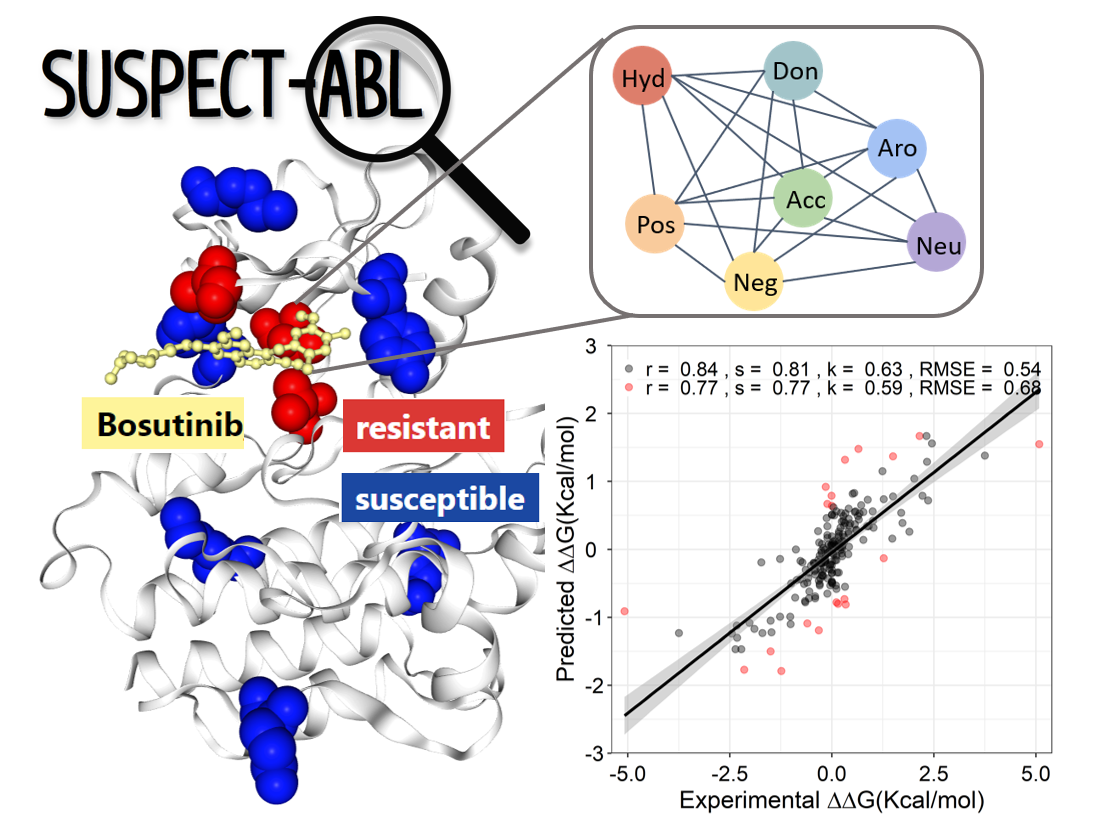
Here we developed a novel, web-based diagnostic tool, SUSPECT-ABL (StrUctural Susceptibility PrEdiCTion for ABeLson 1 kinase), to pre-emptively predict the resistance profiles and binding free-energy changes (ΔΔG) of all possible ABL1 mutations against eight inhibitors with different binding modes. The predictive model was built using supervised learning and leveraged the concept of graph-based signatures to capture not only the structural properties of the protein and ligands, but also the geometric properties within the residual environment. Resistance mutations in ABL1 were successfully identified, achieving a Matthew’s Correlation Coefficient of up to 0.73 and ΔΔGs were predicted with Pearson’s correlation of up to 0.77. Both prediction performances were consistent across our blind tests. Through an in silico saturation mutagenesis, our tool has identified possibly emerging resistance mutations, which offers opportunities for in vivo experimental validation. We believe SUSPECT-ABL is an important tool not just for improving precision medicine efforts, but for facilitating the development of next-generation inhibitors that are less prone to resistance through early consideration of potential resistance mutation hotspots.