About piscesCSM
Running predictions
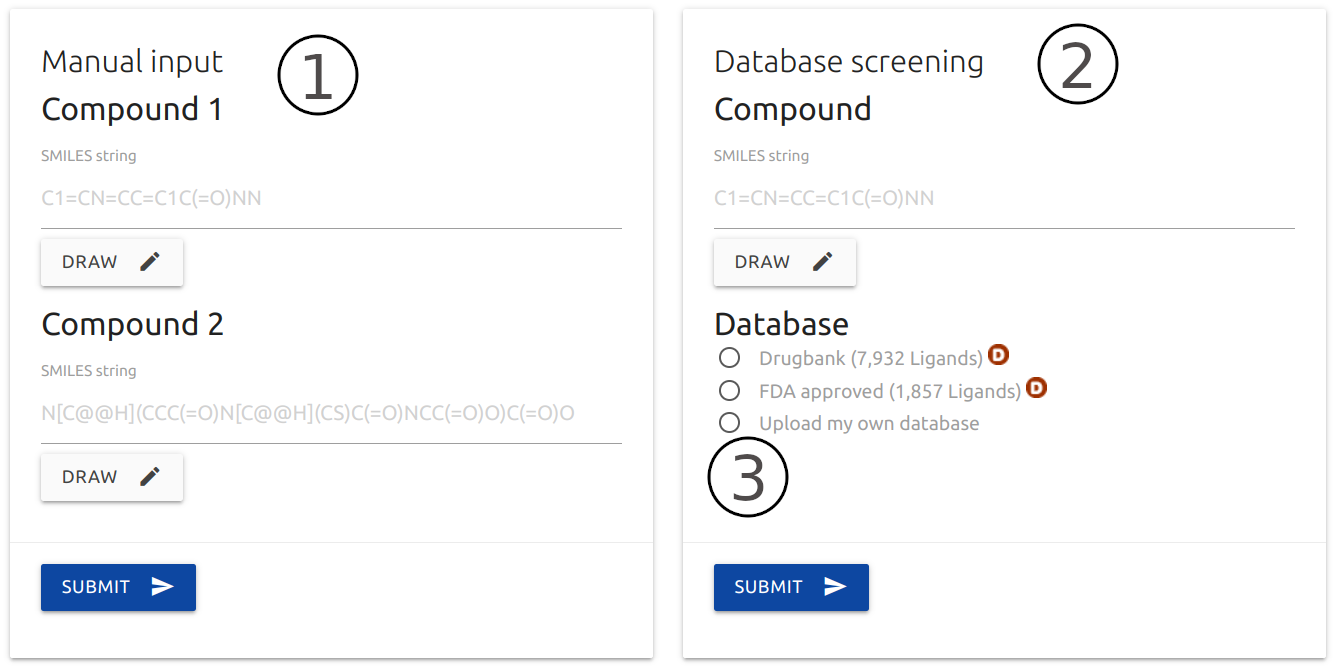
Input

- piscesCSM can be queried with only two small-molecules in SMILES format (1). Users may also use the Kekule.js editor to draw the molecule by clicking the Draw button.
- An option to test a single small-molecule against all compounds on DrugBank or FDA approved drugs is also available (2).
- Alternatively, users may submit their own list of compounds by providing a headless plain text file with a list of SMILES, one per line (3) or
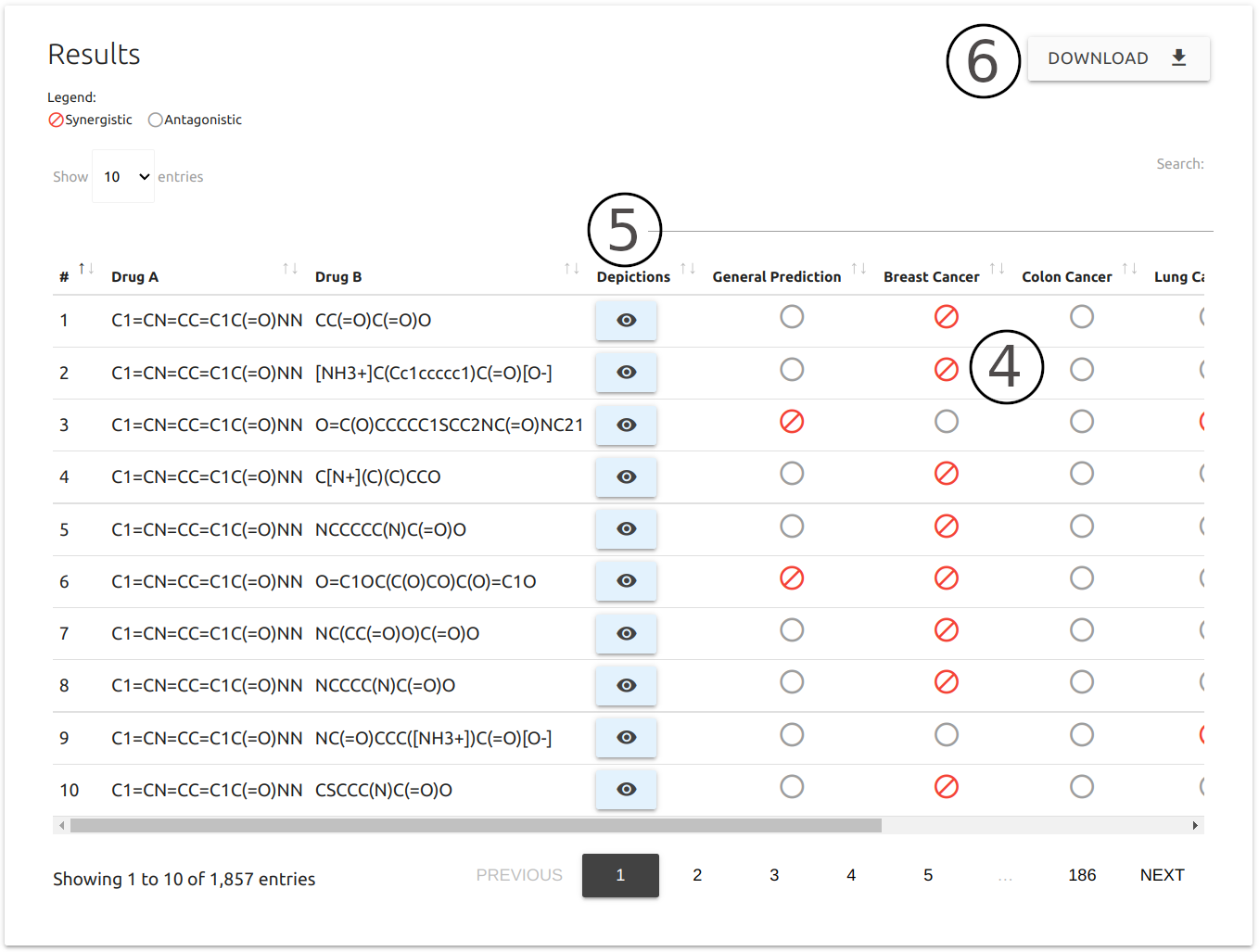
Results

For both input options (Manual and Database Screening), results are summarised as a downloadable table (4)with the following information:
- SMILES for each pair of compounds provided alongside with the predictions, including results for tissue specific predictive models.
- The entire table can be downloaded as a comma-separated values file (CSV) using the button right above the table (6).
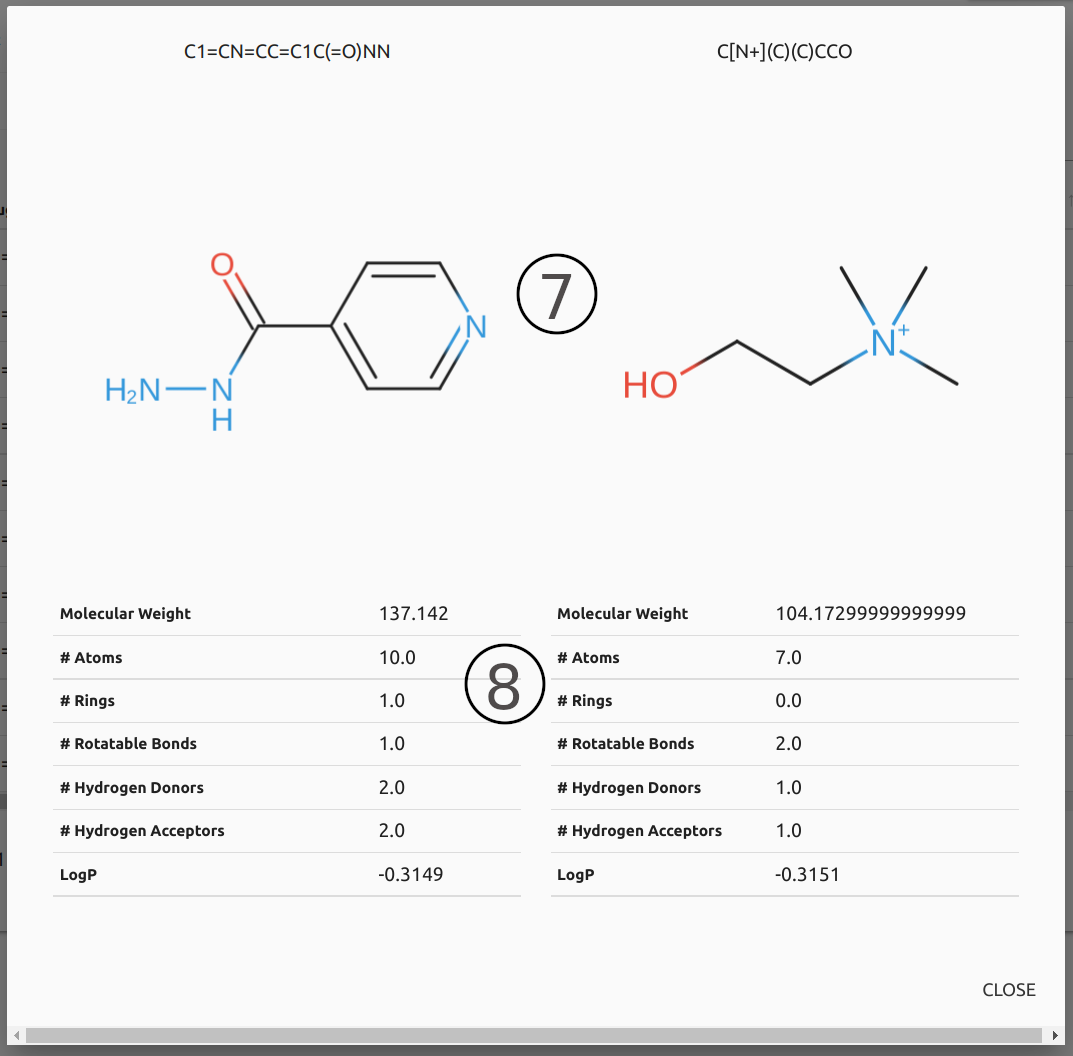
- Users may visualise depiction (7), generated via SmilesDrawer, and general physicochemical properties (8), including Molecular weight, number hydrogen donors and acceptors, and LogP, by clicking on the buttons available in the table.

Contact us
In case you experience any troube using piscesCSM or if you have any suggestions or comments, please do not hesitate in cantacting us either via email or via our Group website.
If your are contacting regarding a job submission, please include details such as input information and the job identifier