Running predictions
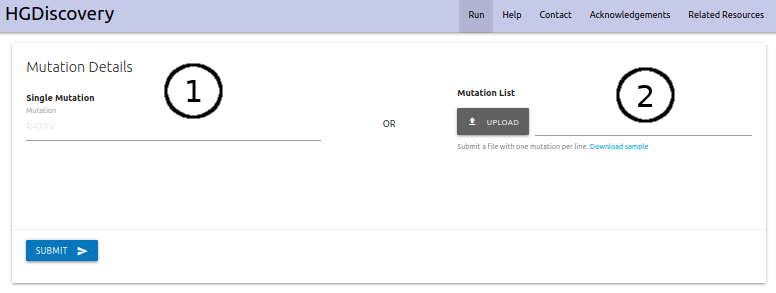
HGDiscovery allows for users to query for a single point mutation or submit alist of mutations to be analysed in batch. For the “Single Mutation” option (1) users are asked to provide the point mutation as a string containing the wild-type reside one-letter code, its corresponding residue number and the mutant residue one-letter code. The “Mutation List” option (2) requires that atext file is submitted with the list of mutations (one per line).
Results
Single Mutation
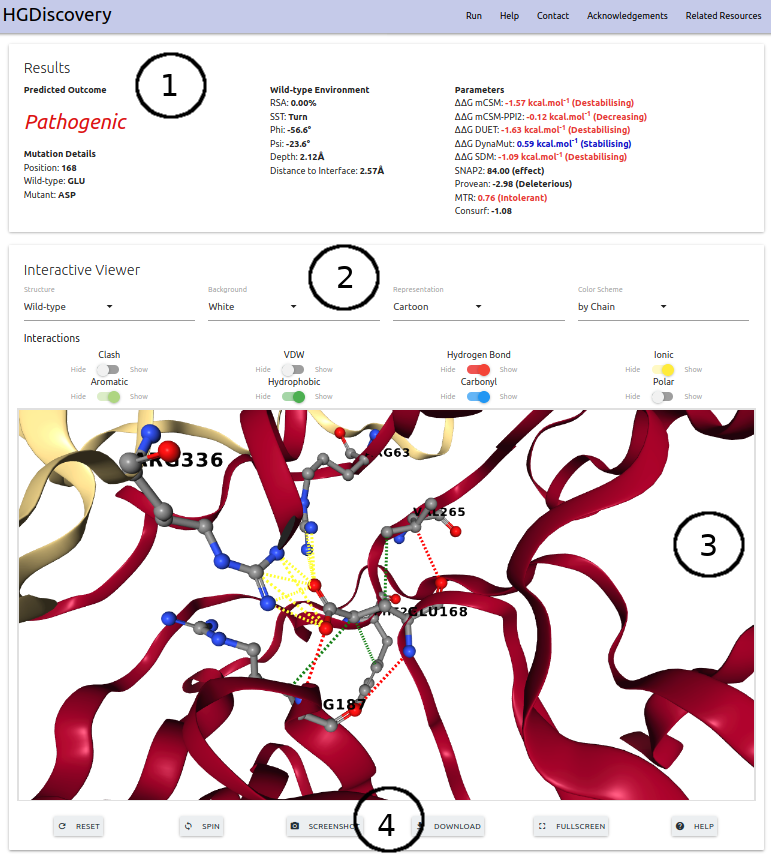
The results page for the “Single Mutation” option displays the predicted outcome on the top alongside with details of the input mutation, wild-typeresidue environment, the variables and scores used by our predictive modeland external links to experimental evidence (when available) (1). An interactive3D (3) shows the molecular contacts generated by Arpeggio for wild-type and mutant structures. Controllers are available for customising the viewer (2). A series of action buttons (4)are also available at the bottom of the 3D viewer.
Mutation List
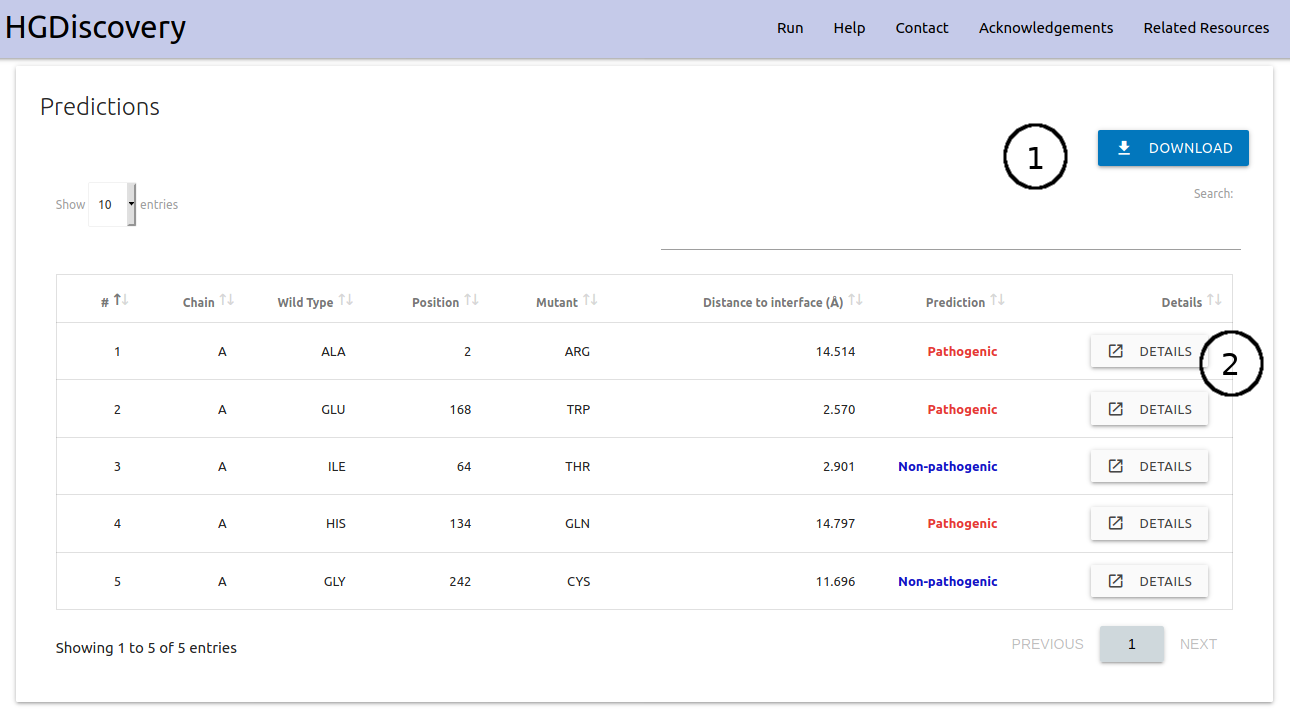
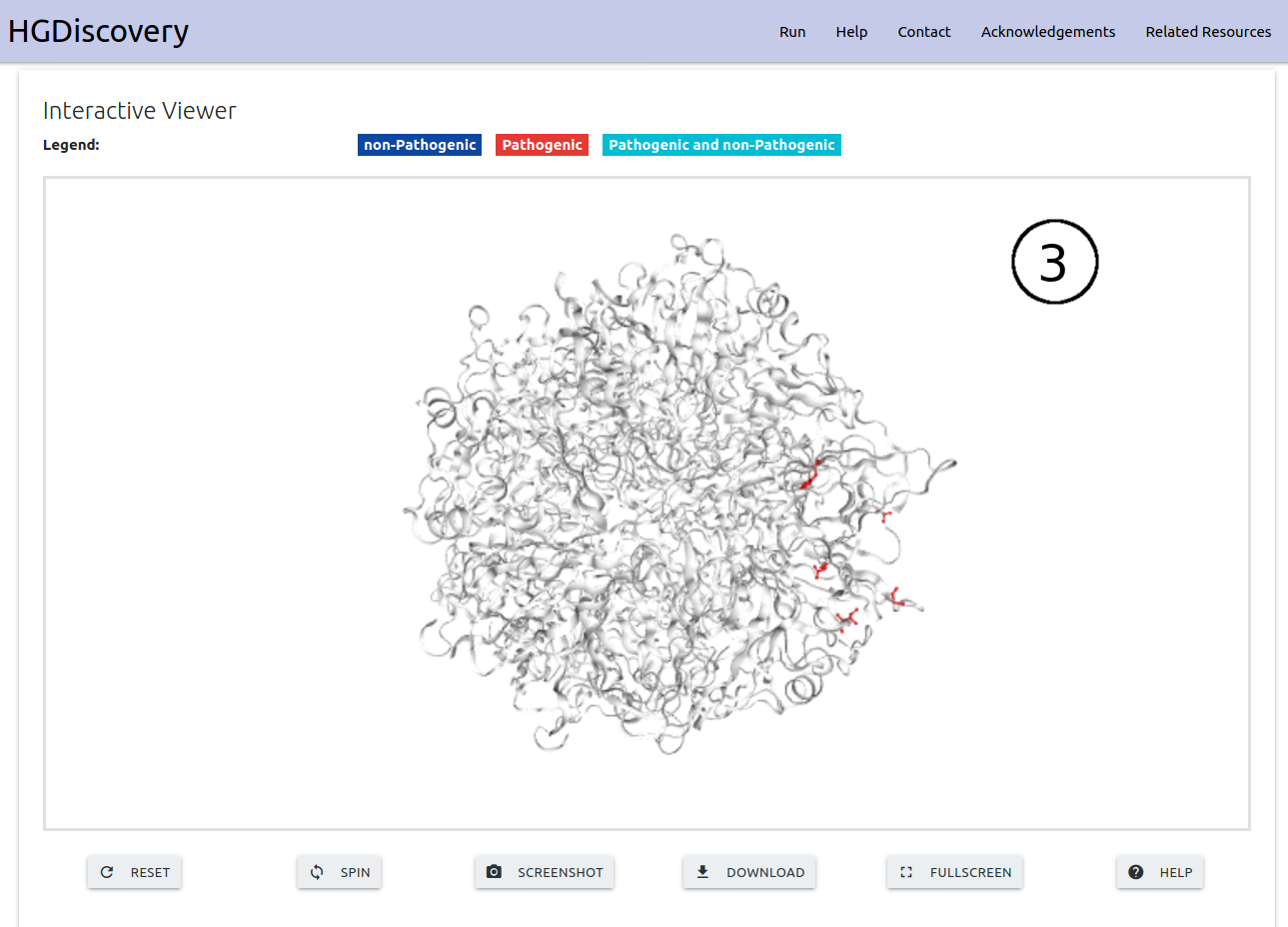
On the “Mutation List” option, the results are displayed as a downloadable table (1). Individual analysis for each variant on the table can be analysed similarly to “Single Mutation” option by clicking the “Details” button (2). An interactive viewer is also shown at the bottom of the page highlighting Pathogenic and Non-pathogenic mutations on the 3D structure (3).
Contact us
In case you experience any trouble using HGDiscovery or if you have any suggestions or comments, please do not hesitate in contacting us via our Group website.
If your are contacting regarding a job submission, please include details such as input information and the job identifier