toxCSM Help Page
About toxCSM
toxCSM is a robust machine learning method that relies on graph-based signatures, molecular descriptors and toxic-active molecular similarities to predict small molecule toxicity profiles. Currently, toxCSM is the most comprehensive method in the literature, encompassing 36 different endpoint properties, which vary from nuclear response to environmental chemical activity.
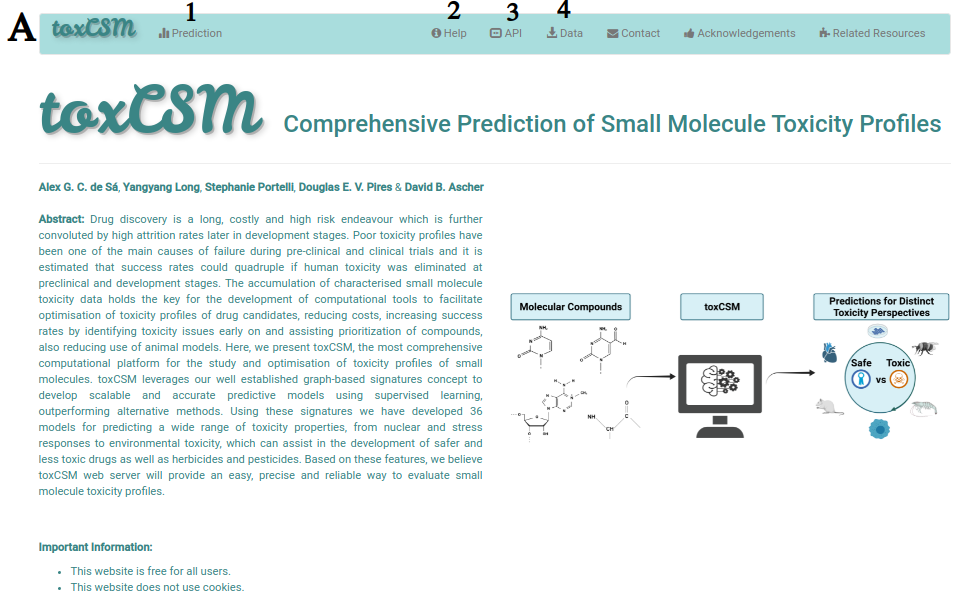
A depicts the main page of toxCSM. Users are directed to the job submission page by clicking on "Predictio" at the top menu (1). Users can also access this help page on "Help" (2), how to use the "API" (3), and all data used in toxCSM experiments by clicking on "Data" at the top menu (4).
Job Submission Page
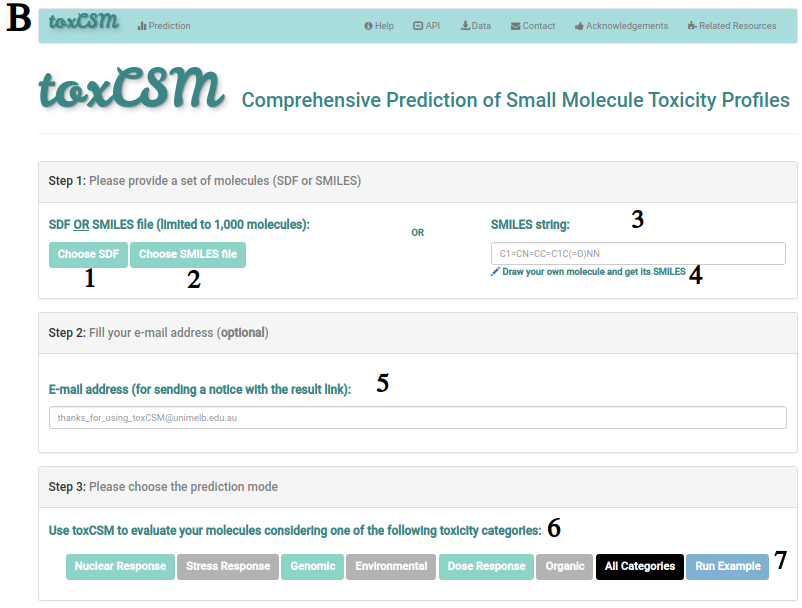
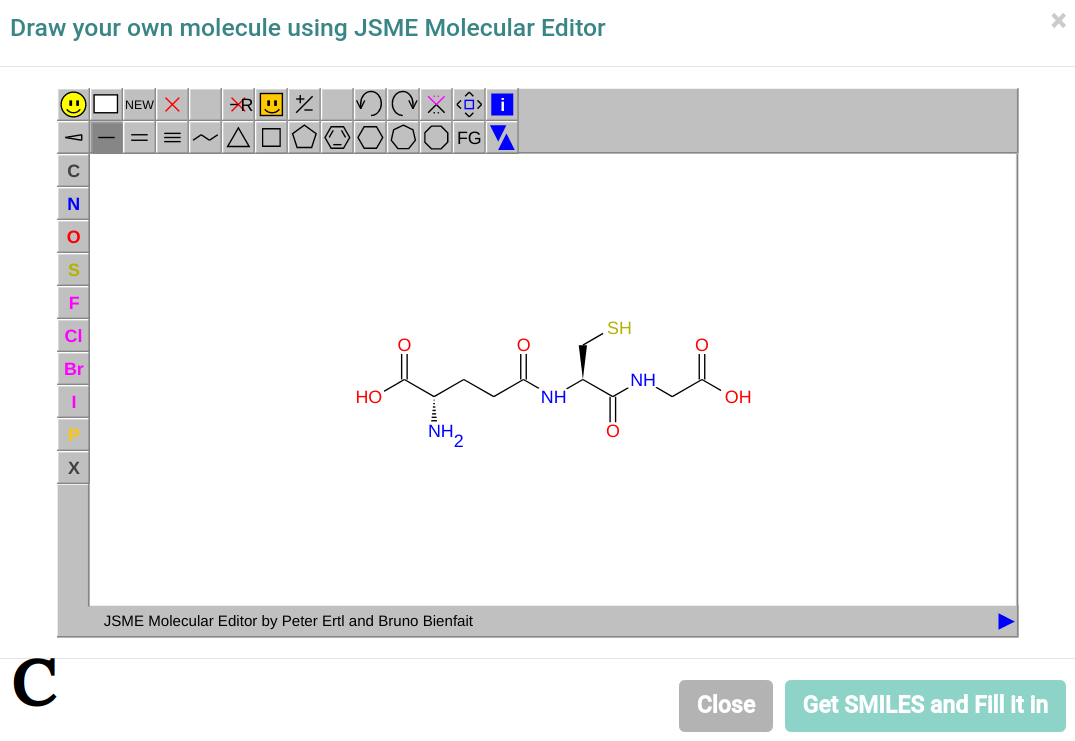
B describes the job submission page, where users can either submit an SDF file (1), a SMILES file (2) or an individual SMILES string (3). For the SMILES string, users can also draw their molecule using JSME molecular editor and retrieve its SMILES, as shown at C. It is worth noting when the user clicks on "Get SMILES and Fill It In" at C, the SMILES string (3) at B will be filled with the respective SMILES of the drawn molecule.
Besides, if users want to, they can provide an email address (5) at the submission form.
After these steps, the user can choose among several toxicity categories to run predictions on (6), including Nuclear Response, Stress Response, Genomic, Environmental, Dose Response, Organic and All Categories. Users can also run an example by clicking in "Run Example" (7).
Waiting Page
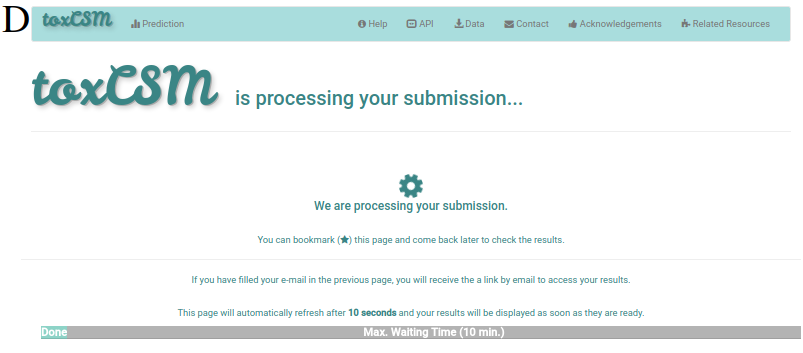
When the user clicks on any perspective category option at B (6), he/she will be redirected to a waiting page while the job is being run (D). If the email address was filled on the previous page, the user can wait for an email with the link to access the results.
Error Page
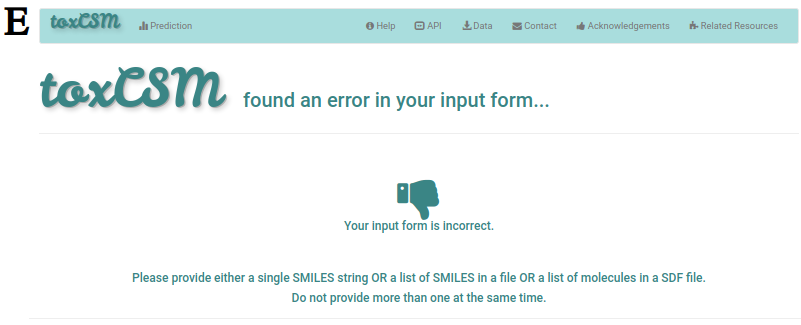
If anything goes wrong in the submission or job processing, the user will visualise an error page such as the one presented at E. If the email address was filled on the previous page and the job already started its processing, the user will also receive an email mentioning the occurred issue.
Results Page
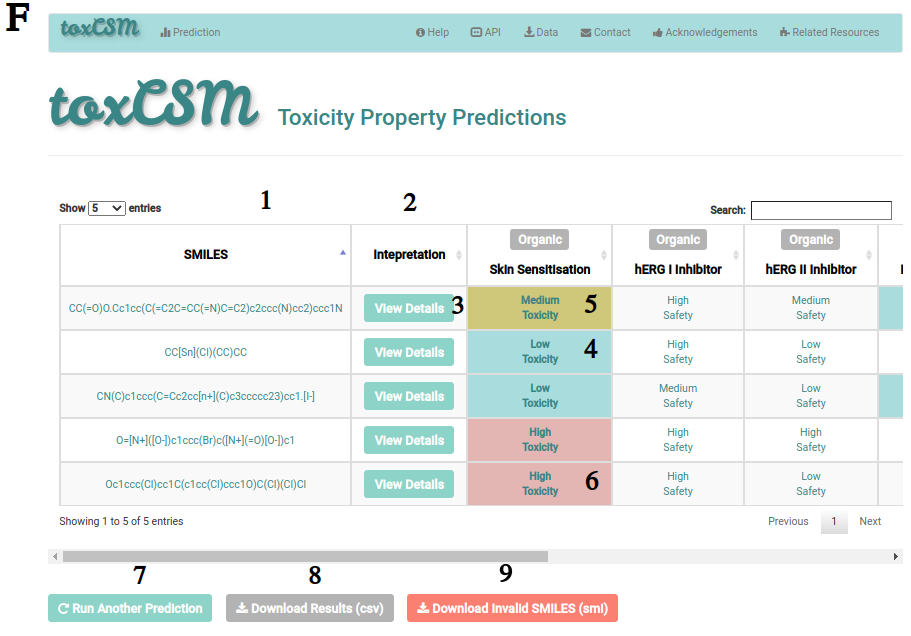
F describes the result page for five molecules. It includes the SMILES of the molecule (1), a set of interpretation buttons (2, 3) and the predictions for the selected toxicity categories (4, 5, 6). Classification predictions are presented in terms of high, low and medium toxicity or safety as exemplified at F. Regression predictions, in turn, are presented as real-valued numbers. In classification tasks, compounds with high (6), medium (5) and low (4) toxic profiles are coloured in red, mustard and cyan, respectively. On the other hand, safe compounds are kept in grey or white. Users can also run another prediction (7), download the results into a comma-separated value (csv) file (8) and, if any SMILES was considered as invalid, the user can download them within a (smi) file (9). The button presented in (9) will appear only when invalid compounds are found.
Analysis Waiting Page
If the user clicks at any button "View Details" (3) at F, users will be redirected to a waiting page again while the analysis of that particular molecule is being performed. This waiting page is presented at G.
Analysis Page
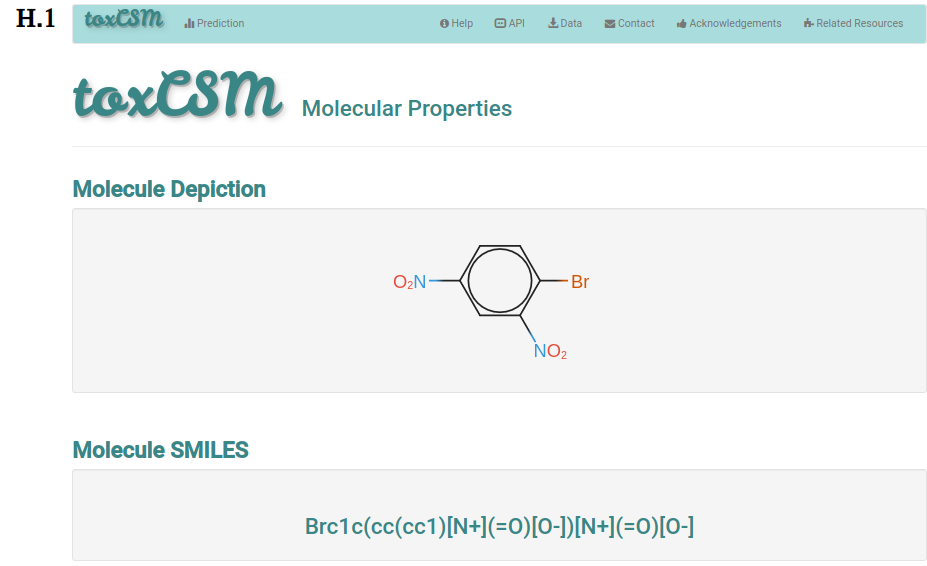
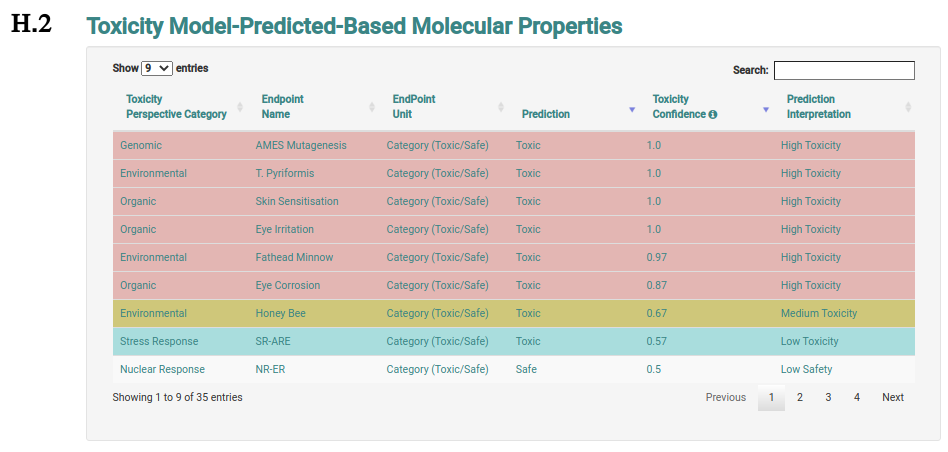
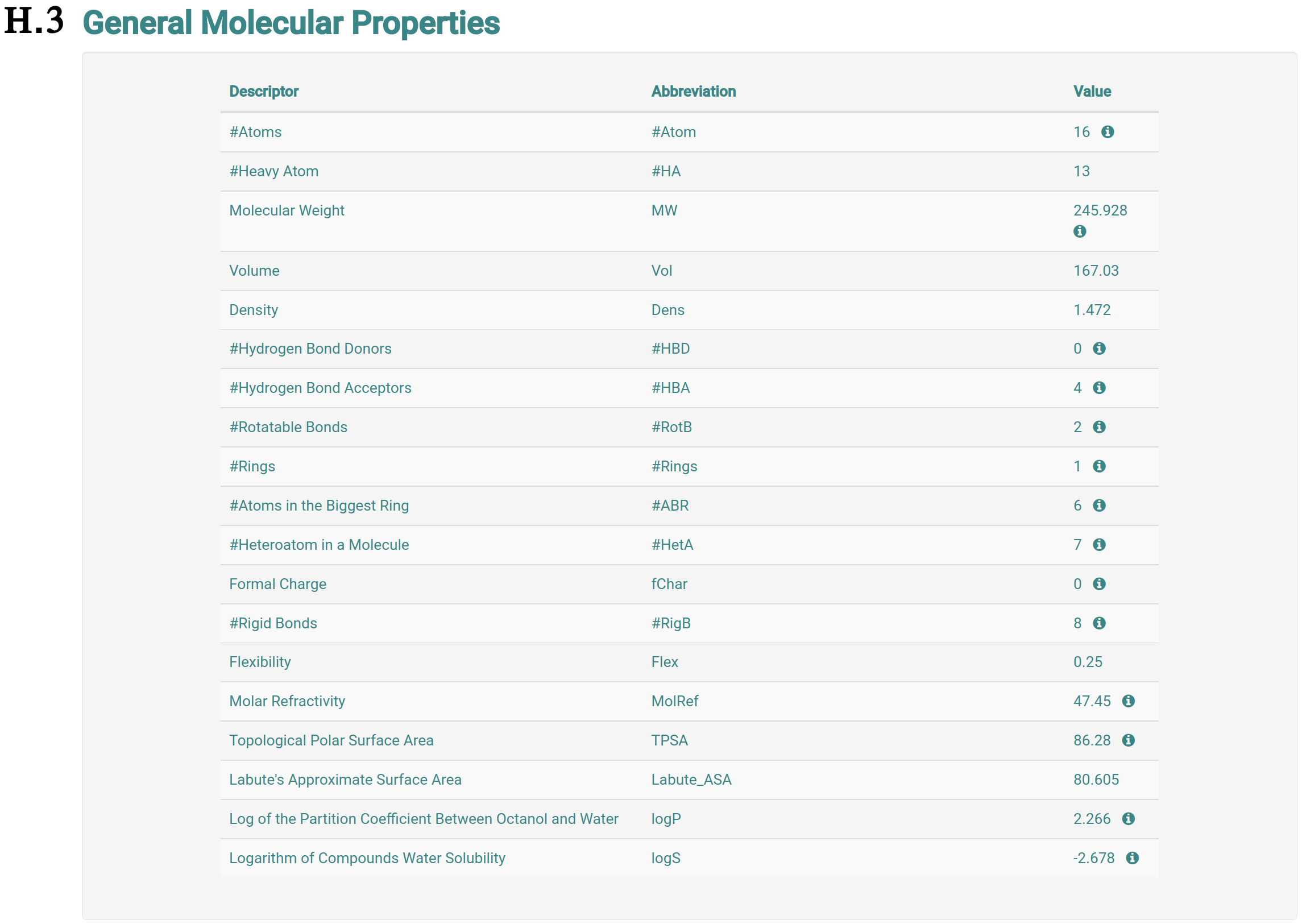
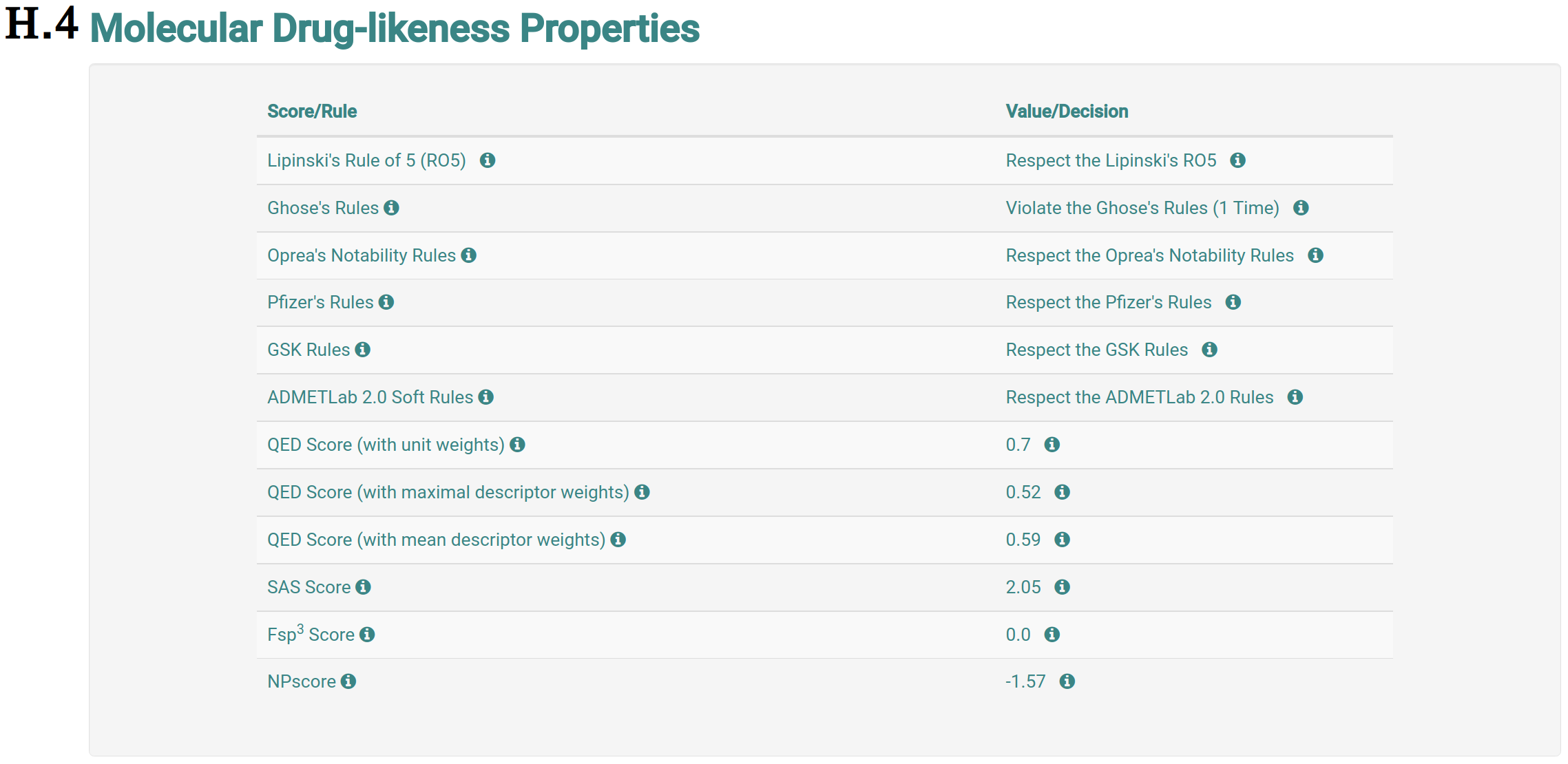
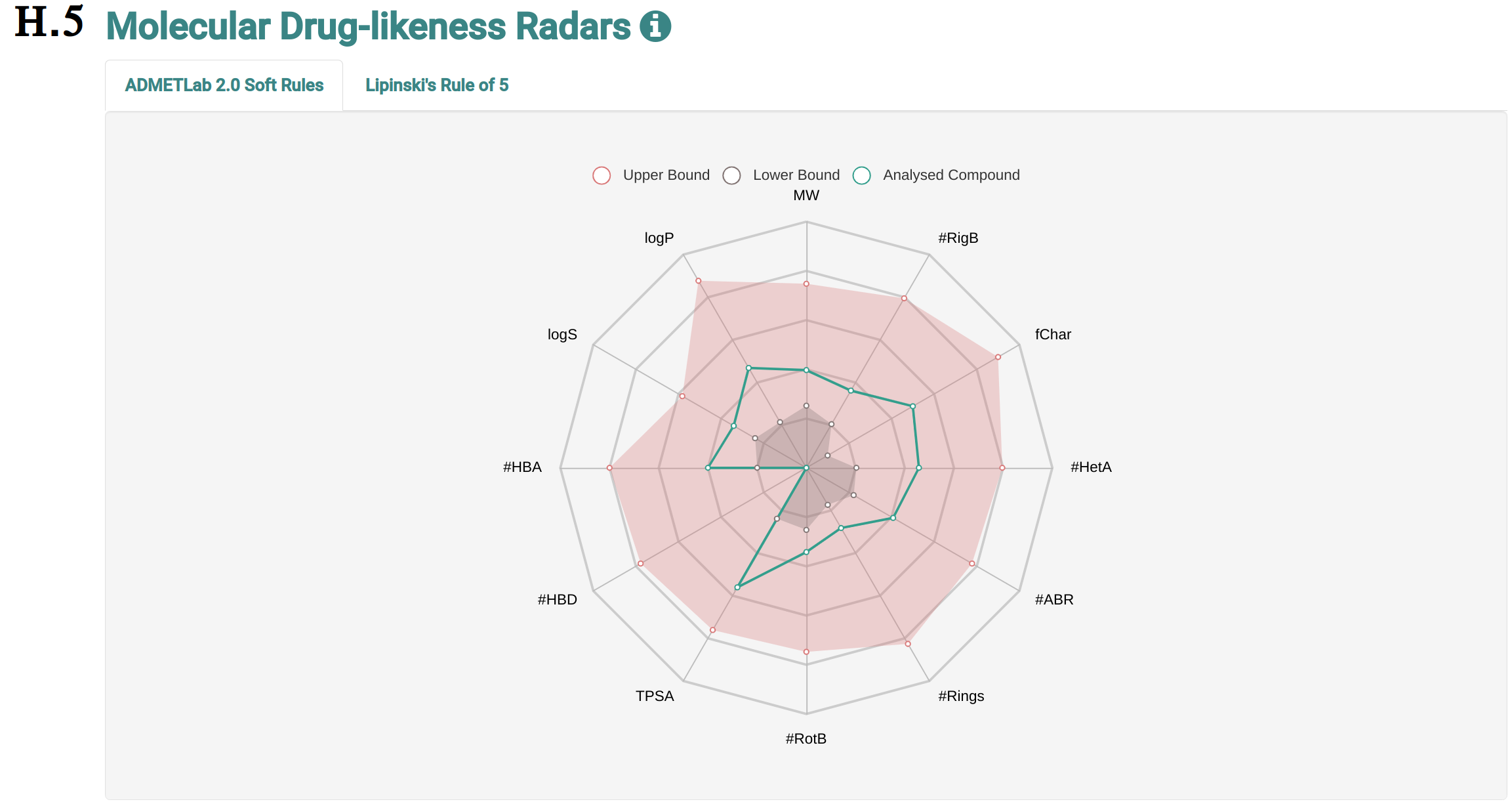
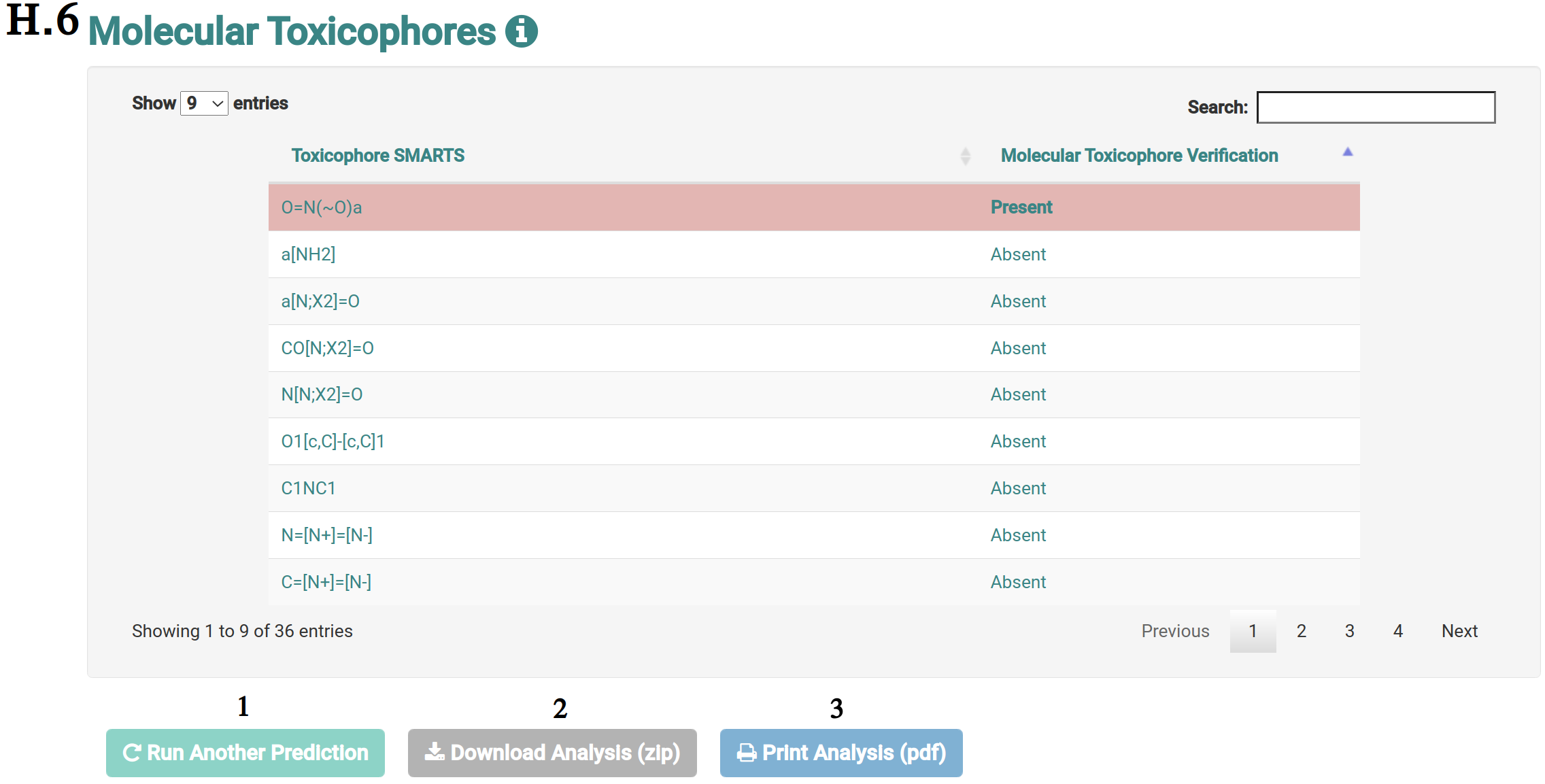
The analysis page shows information of the molecule the user requested more details about. This page is presented from H.1 to H.5, comprehending the molecule depiction and its respective SMILES (H.1), the toxicity properties (model predictions) sorted by highest toxic confidence level (H.2) -- if the molecule is considered to be toxic in a particular endpoint model, the color of the model’s row is also highlighted in red, mustard and cyan to indicate high, medium and low toxicity profiles, respectively, the general molecular properties (H.3), the molecular drug-likeness properties (H.4), the molecular drug-likeness radar plots (H.5), the analysis of presence (highlighted in red) or absence of 36 toxicophores (H.6) -- toxicophores are sorted based on their presence in the analysed molecule.
Users can also be redirected to run another prediction (H.6/1), download the analysis of the molecule in a zip file (H.6/2) -- which encompases the tables at H.2, H.3, H.4 and H.6 or print the analysis page (H.6/3).
In the whole page H, tooltips are used to help the interpretation of the analysis.
Data Page
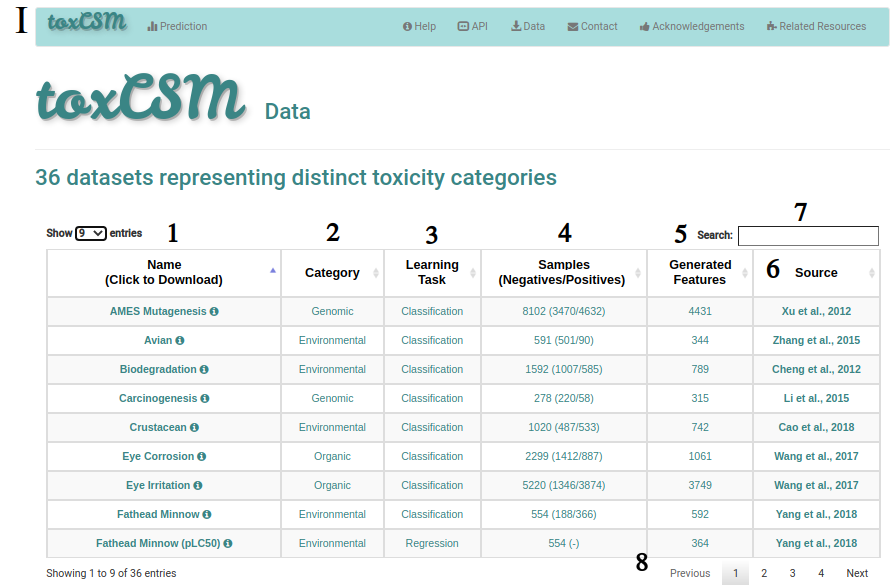
If the user clicks on the data page at the top menu, he/she will be redirected to the data page I, where all the endpoints used to train, validate and test toxCSM predictive models. Users can access the name of the endpoint data (1) and download it by clicking on their names. In addition, information about the perspective category (2), machine learning task (3), number of samples (4), number of generated features (5), and source (6) are available. Users can also search (7) for the respective endpoint data and browse across them (8).
In the whole page I, tooltips are available to assist the user.
Contact Us
In case you experience any troube using toxCSM or if you have any suggestions or comments, please do not hesitate in cantacting us either via email or via our Group website.
If your are contacting regarding a job submission, please include details such as input information and the job identifier