PRIMITI Help
Index
About PRIMITI

PRedictive model for Identification of novel MIRNA-Target mRNA Interaction (PRIMITI) is a machine learning model that utilizes diverse CLIP-seq and expression data to predict novel miRNA-target mRNA interactions. The main focus of PRIMITI model is a miRNA-induced post-transcriptional repression caused by canonical binding between miRNAs and target sites in 3'-UTR of protein-coding transcripts. That is because the canonical binding in 3'-UTR is considered as a functional site type that is more likely to lead to transcriptional repression.
PRIMITI utilizes 22 distinct features to describe physical miRNA-target site binding/duplex pairing and 4 features to describe miRNA-target mRNA repression.
Marked sections in the above toolbox, include:
A depicts the main page of PRIMITI. Users are directed to the job submission page by clicking on Prediction (1)
page at the top menu. Help (2) also available to instruct the users how to use PRIMITI.
Data (3) provides the user with all data used in PRIMITI training, (cross-validation) and evaluation.
Job Submission Page

In the Job submission page (B), users need to provide identifiers for miRNAs and candidate target messenger RNA (mRNA). Users may provide the information as a file (1/3) or as a string (2/4).
The example files for both miRNA and protein-coding transcript (linked to mRNA) can be downloaded from the buttons.
For transcripts, Ensembl transcript (ENST) form for inputs are highly preferred, as our model makes predictions using the information of the transcript.
Nevertheless, our web server still supports several gene ID formats, andusers can instead provide gene identifiers in place of transcripts.
All the protein-coding transcripts associated with a given gene will be examined.
For instance, If ENSG00000141510 (TP53) is provided, all the protein-coding transcripts for TP53 will be analysed in the PRIMITI model.
The list of supported input format for transcript/gene are as following:
- 1) Ensemble Transcript : ENST00000340080
- 2) Ensemble Gene : ENSG00000141510
- 3) NCBI RefSeq Transcript : NM_001003806, NP_001263195
- 4) NCBI Gene name : SIDT2, TP53
- 5) NCBI Gene ID : NCBIGeneID:7157, NCBIGeneID:1956
- 6) MIM Gene ID : MIM:165280, MIM:613919
- 7) HGNC Gene ID : HGNC:11547, HGNC:46137
Please note that the analysis request that contains a gene identifier will take longer to process when compared to those with only containing transcripts.
If users are interested in the analysis of both gene and transcript, we recommend submitting the jobs individually.
An email address can be also provided by the user (in an optional form) (5), so that the user can be notified with an access link when the analysis is finished.
Notably, our webserver only accept the number of candidate interactions (number of miRNAs x number of transcripts) up to 30,000. If the user’s data is larger than the limitation, please consider submitting them separately. In a case that gene identifiers are provided, the number of candidate interactions will be multiplied by 3. (For example, 1 miRNA and 1 gene = 3 candidates interactions)
Waiting Page
After submitting the analysis, the user will be redirected to a waiting page (C). If the email address is filled in (B), user will receive the link to the result page (D) in the email.
Result Page - Main

The Result Page (D) provides the information of predicted miRNA-target mRNA. Pairs of miRNA and transcript (representing mRNAs)
that contain at least one canonical binding site are shown in the table of this page.
The identifiers for miRNAs (1) and input transcripts/genes (2) are provided along with Ensemble Transcript (ENST) (3)
and Ensemble Gene (ENSG) (4).
If a miRNA-transcript pair is predicted to exhibit a post-transcriptional repression, the Interaction (5) will be shown as "Yes", otherwise "No".
The confidence score for each pair is provided in the column Interaction Confidence (6). To allow a high-confidence prediction, a cut-off of 0.9 was set for predicting the repression.
The numbers of all candidates canonical target sites are shown in No. of target sites (7).
If users are interested in more detailed information regarding the physical binding in each target site, please click on the button in More details (8) for each miRNA-transcript pair. PRIMITI web server will redirect you to the Further information page (E).
Three bottons in D allow users to run another prediction (9) as well as to download results of miRNA-target mRNA repression (10),
results of miRNA-target site binding for all miRNA-target mRNA pairs (11) and an error log (12).
Result Page - Further information

Result Page - Further Information (E) shows the detailed information of all physical miRNA-target site binding in a pair of miRNA-transcript. In the upper table,
miRNA ID (1), Provided ID, Transcript ENST (2), Gene ENSG (3) are provided with a predicted interaction and respective confidence score for that pair. For further detail regarding the miRNA, transcript, and gene,
clicking the link will direct users to miRBase (https://www.mirbase.org/) and Ensembl database (https://www.ensembl.org).
The next table provides the following information for each target site (4): Binding status (5), Binding Confidence score (6), Position in terms of 3'-UTR (7) or cDNA (8),
and the type of canonical binding (6-mer, 7-mer-A1, 7-mer-m8, and 8-mer) (9).
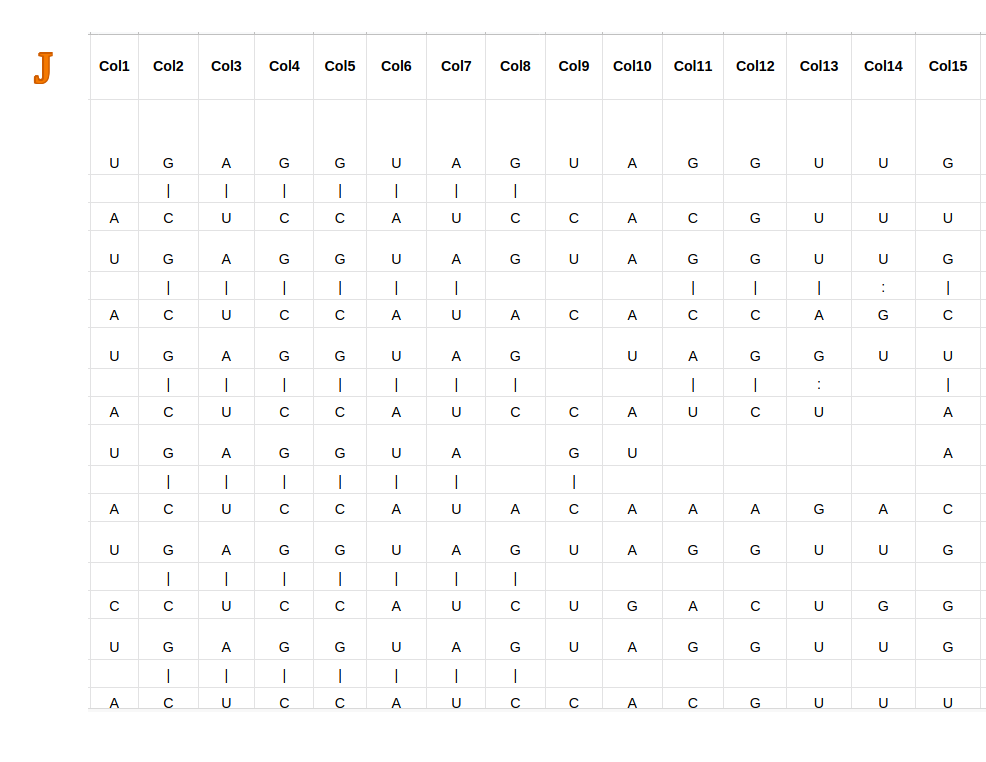
Predicted miRNA-target site duplex structure (10) is shown as a picture. The upper sequence is miRNA sequence (5' to 3') and the bottom is mRNA sequence (3' to 5').
Across the sequence, the predictions of the base pairwise interactions are shown. One line represents one base pairwise.
Additionally, as the presence of adenine (A) nucleobase in the first position of mRNA (3’) strongly correlates with a significantly higher miRNA-mRNA duplex stability,
Adenine nucleotide will be depicted in brown if it is present at the first position.
Error Page
The Error Page (F) is shown when your input is incorrect. Please check the example files for an input format.
Common problems are 1) Wrong format in input files 2) Swap miRNA and transcript input files
If you have any inquiry, please do not hesitate to contact us via our Group website.
Downloadable results
The example of downloadable results
- Predicted miRNA-target mRNA repressions (G)
- Predicted miRNA-target site interactions (H/I)
- Error log
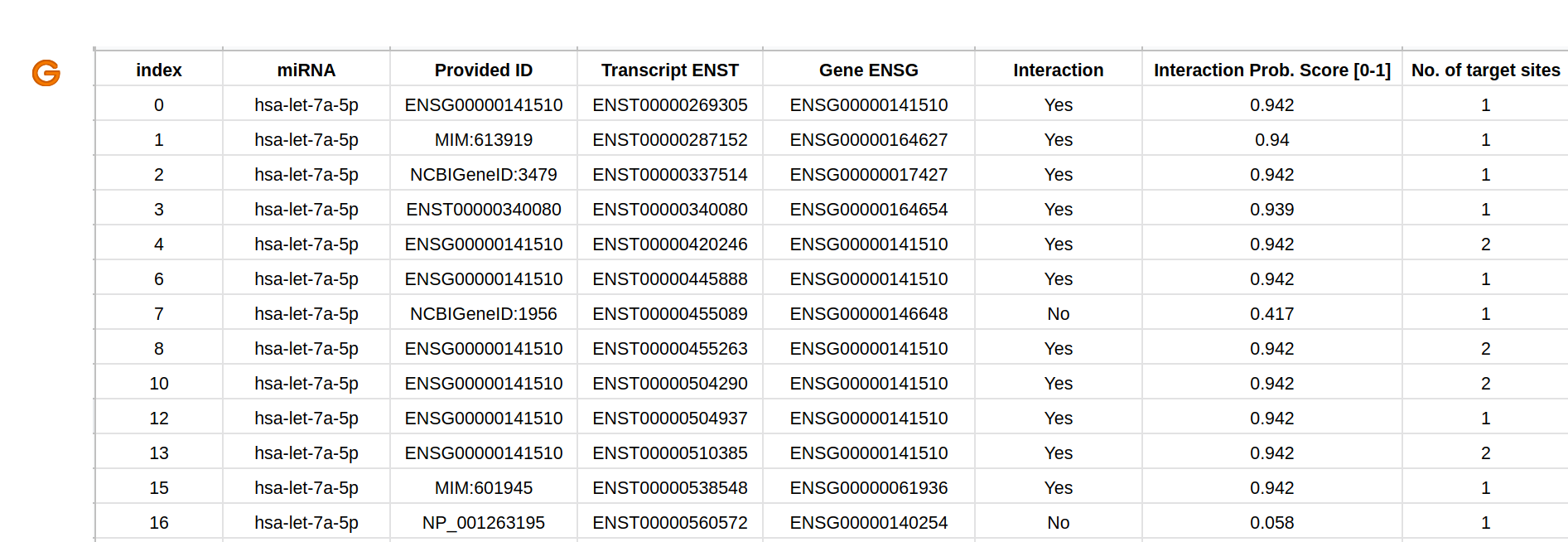
The predicted miRNA-target mRNA repressions (G) can be retrieved from a button in Result Page - Main (D-10) as a csv file.
The provided information is the same as in result table in (D). It contains identifiers for miRNA, Provided ID, Transcript ENST, Gene ENSG, Interaction status
(Yes: post-transcriptional repression, No: no repression), Confidence score, and Number of target sites.
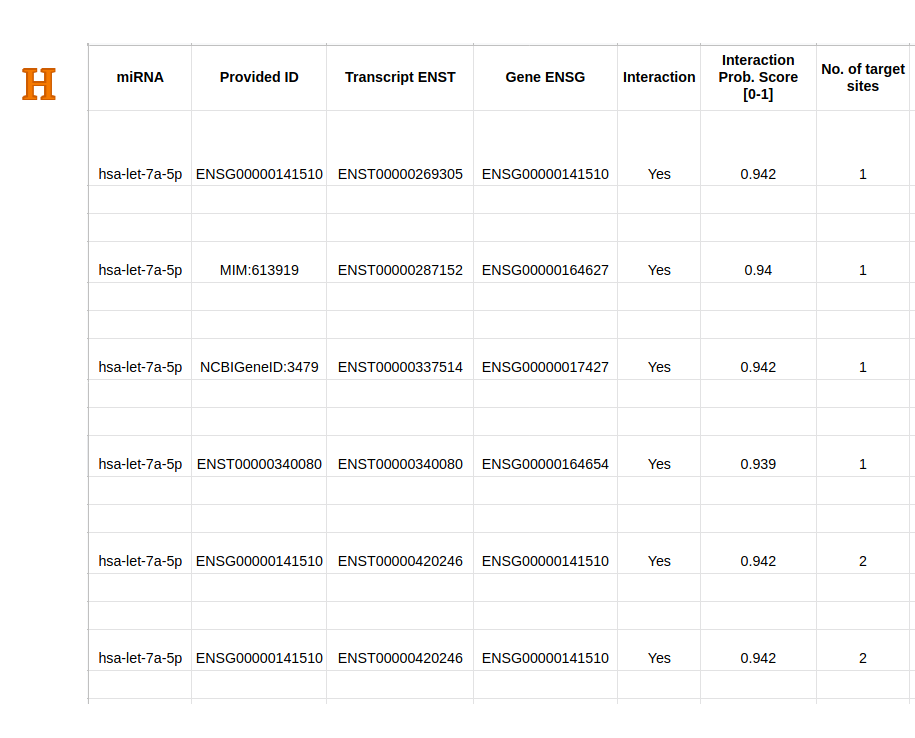
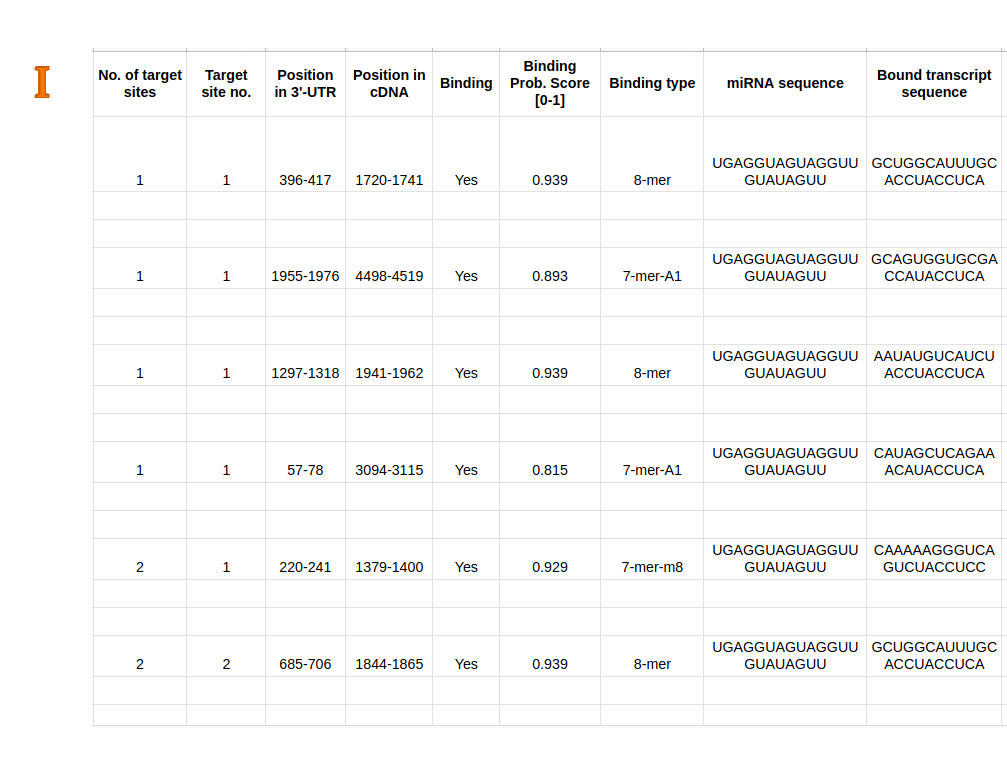
Predicted miRNA-target site interactions (H/I/J) can be retrieved using the dedicated download button in Result Page - Main (D-11) as a csv file.
The provided information contain predicted miRNA-target mRNA repression (same as in (D)), and data in each target site, including the Predicted binding status
(Yes: miRNA can bind with target site, No: they are not binding), Binding confidence (range from No (0) to Yes (1)), Binding type (6-mer, 7-mer-A1, 7-mer-m8, and 8-mer),
mRNA sequence, Bound transcript sequence, and Predicted duplex structure calculated by IntaRNA.
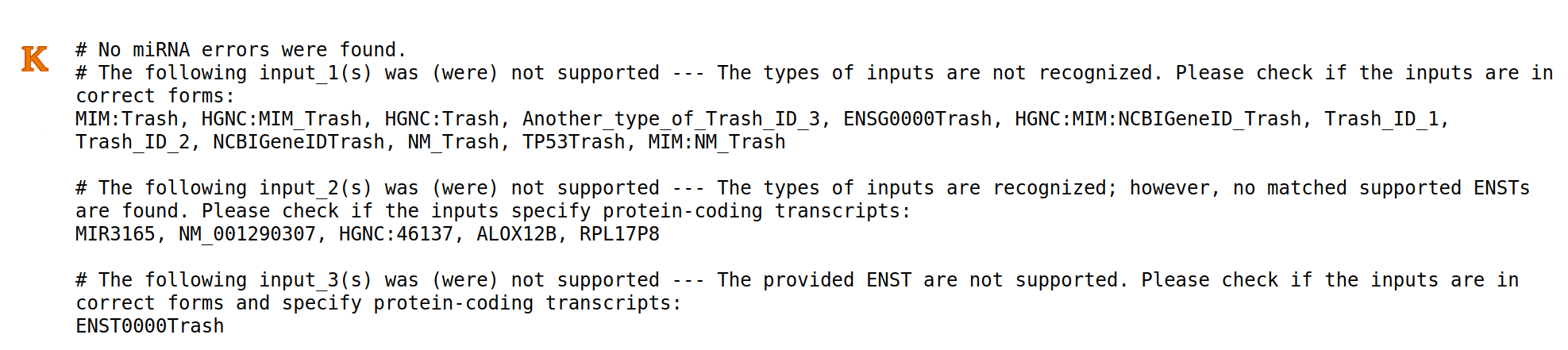
An error log (K) can be downloaded using the dedicated button in Result Page - Main (D-12). The error entries and related causes will be reported in this file.
Please note that the analysis submission will still be done even though some error entries are found. That is, the error entries will be ignored and the analysis will be accomplished with the remaining correct entries.
To illustrate, if users provide 100 transcripts, in which 5 of them are not correct, the analysis will be performed with 95 transcripts.
4 Type of errors will be reported.
- 1) Errors in miRNAs: most likely occur from incorrect miRNA format.
- 2) Errors in transcript/gene inputs: The type of the inputs is not recognizable. It is more likely that the inputs are in incorrect forms. For example, Trash_ID_1 and Trash_ID_2
- 3) Errors in transcript/gene inputs: The type of inputs is recognizable, but it does not specify any protein-coding transcripts. For example, RPL17PB specifies a pseudogene, which can be found in the database, but not supported in the model.
- 4) Errors in ENST inputs: The provided ENST are not supported. It can be caused by 1) incorrect forms of ENST (For example, ENST0000Trash) 2) Not a protein-coding transcript.