pdCSM-PPI using graph-based signatures to identify protein-protein interaction inhibitors
Carlos H.M. Rodrigues, Douglas E.V. Pires & David B. Ascher
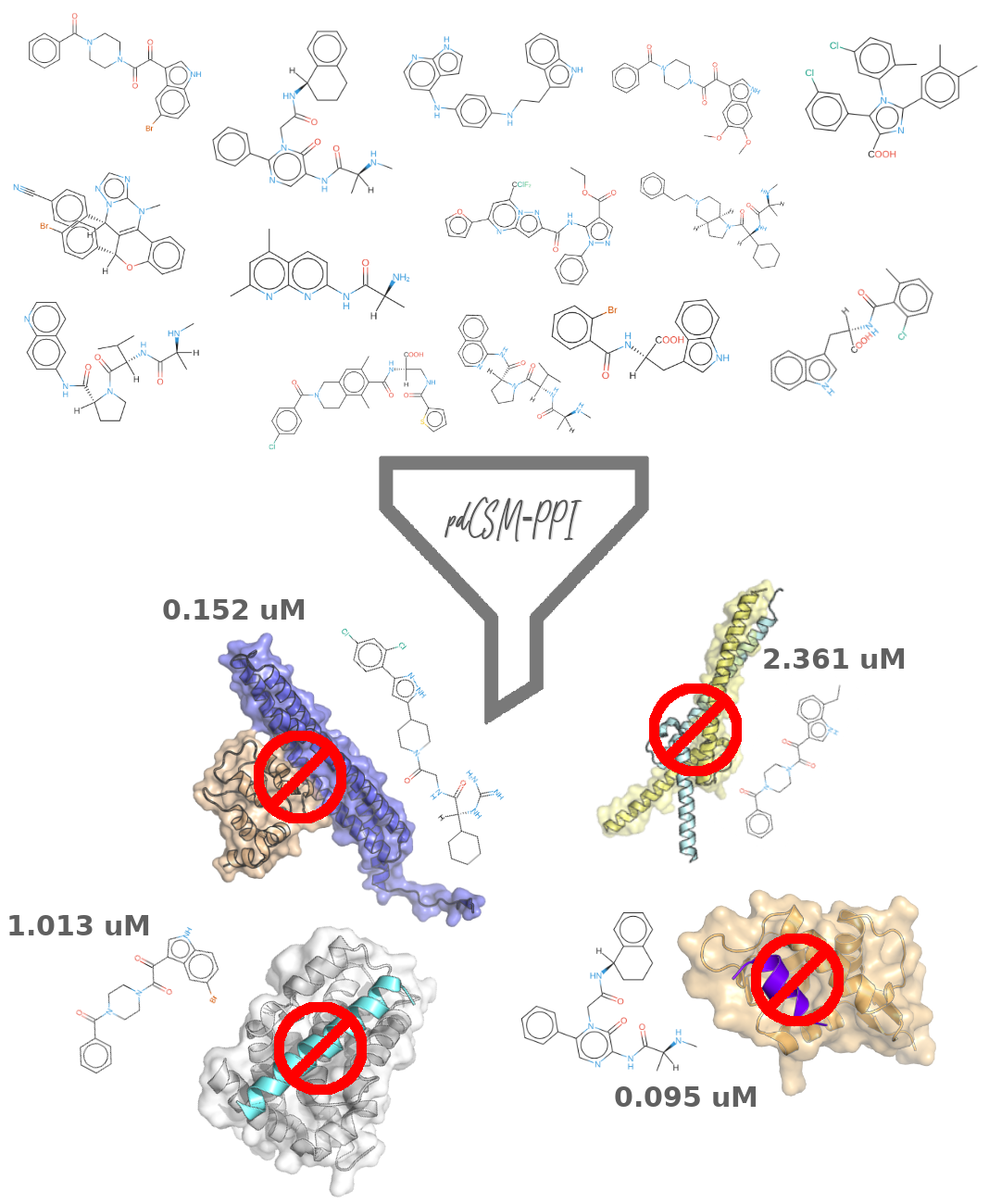
Abstract: Protein-protein interactions are promising sites for development of selective drugs, however they have generally been viewed as challenging targets. Molecules targeting protein-protein interactions tend to be larger and more lipophilic than other drug-like molecules, mimicking the properties of interacting interfaces. Here we propose a machine learning approach that uses a graph-based representation of small molecules to help guide identification of modulating protein-protein interactions via inhibition, pdCSM-PPI. This approach was applied to 23 different PPI targets. We developed interaction-specific models that were able to accurately identify active compounds achieving MCC and F1 scores up to 1, and Pearson’s correlations up to 0.87, outperforming previous approaches. Using insights from these individual models, we developed a generic protein-protein interaction modulator predictive model, which accurately predicted IC50 with a Pearson’s correlation of 0.64 on a low redundancy blind test. Importantly, we were able to accurately identify active from inactive compounds, achieving an AUC of 0.77, sensitivity and specificity of 76% and 78%, respectively. We believe pdCSM-PPI will be an important tool to help guide more efficient screening of new PPI inhibitors and have made it freely available as an easy-to-use web server and API.
searchRun prediction