pdCSM-cancer: Help Page
How to use pdCSM-cancer
About pdCSM-cancer
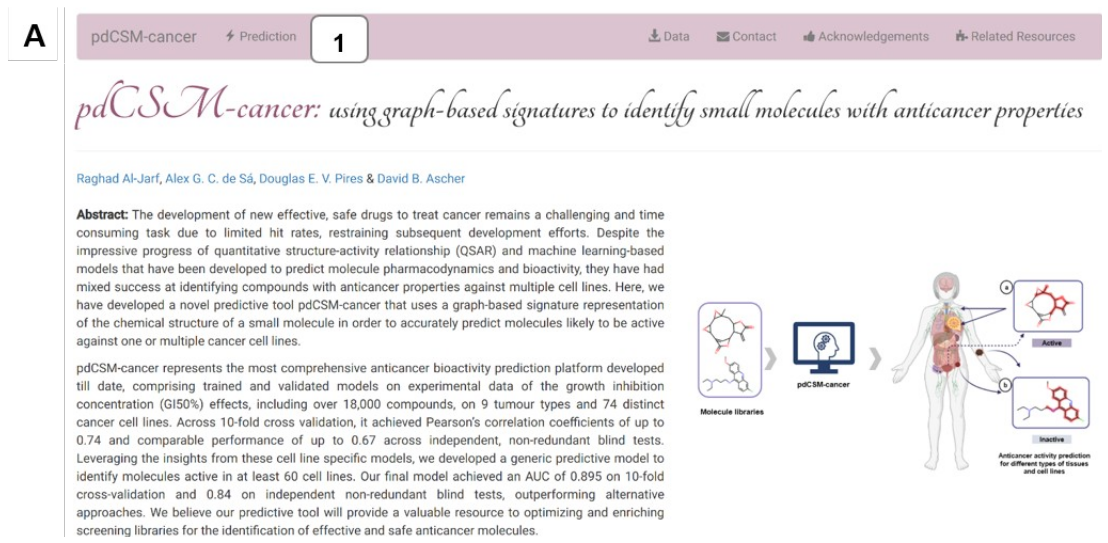
pdCSM-cancer is a machine learning platform that uses a graph-based signature representation of the chemical structure of a small molecule to accurately predict molecules likely to be active against one or multiple cancer cell lines, as well as pharmacodynamics properties. The platform consists of 74 regression models and a general classification model. These models were trained and tested on different experimental data sets of molecules with anticancer properties, tested against nine distinct tissue(tumor) types, including Breast, central nervous system, Colon, Leukemia, Prostate, Renal, Lung, Melanoma, and Ovarian.
(A) Depicts the main page of pdCSM-cancer. Users are directed to the submission page by clicking on “Prediction” at the top menu (1).
Submission Page
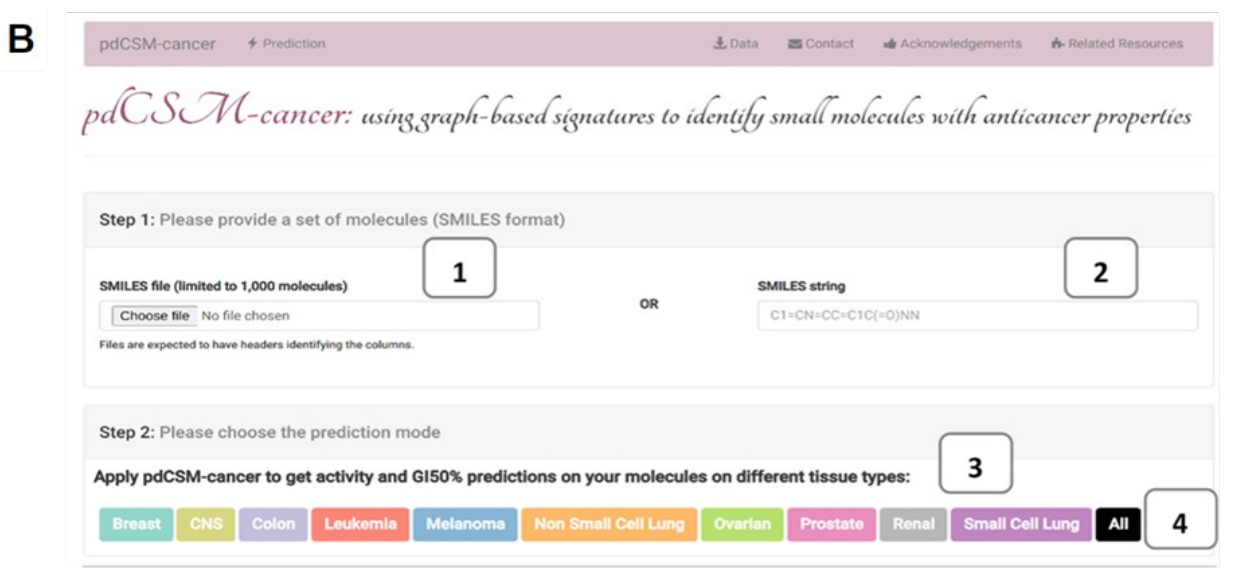
(B) represents the submission page. Users can either submit a set of compounds as a SMILES file (1) or an individual compound as a SMILES string (2). Users have the options to either choose different prediction modes according to their tissue of interest (3) or they can choose to run all tissues (4).
Results Page
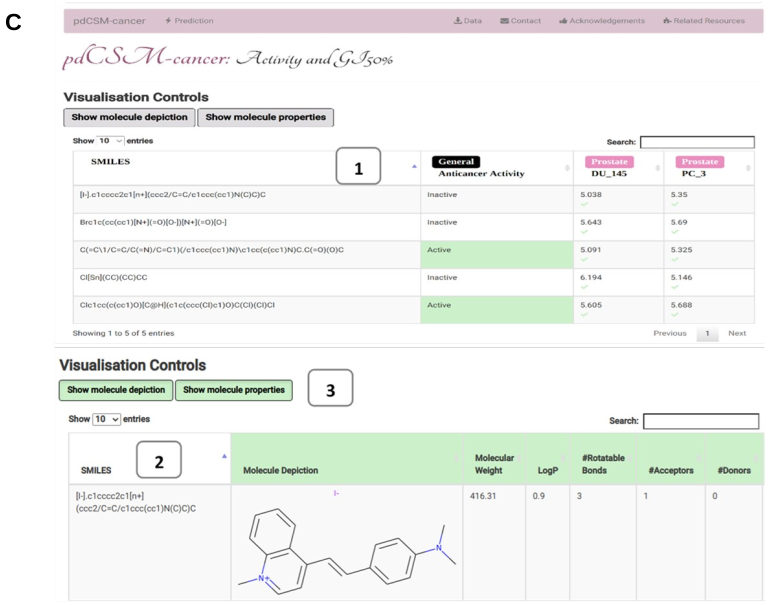
After choosing the prediction mode of interest, users will be redirected to a results page (C) where predictions for all 74 cancer cell lines (10 tissue types) specific models, anticancer activity (GI50%), general anticancer model (1), physiochemical properties and molecular depiction are presented in tabular format (2). Users have the options to either show or hide the molecule properties and depiction (3).
Contact Us
In case you experience any troube using pdCSM-cancer or if you have any suggestions or comments, please do not hesitate in cantacting us either via email or via our Group website.
If your are contacting regarding a job submission, please include details such as input information and the job identifier