MTR3D-AF2: Expanding the Coverage of MTR3D Scores Across the Human Proteome Using AlphaFold2
Aaron Kovacs, Stephanie Portelli, Michael Silk, Carlos H. M. Rodrigues & David B. Ascher
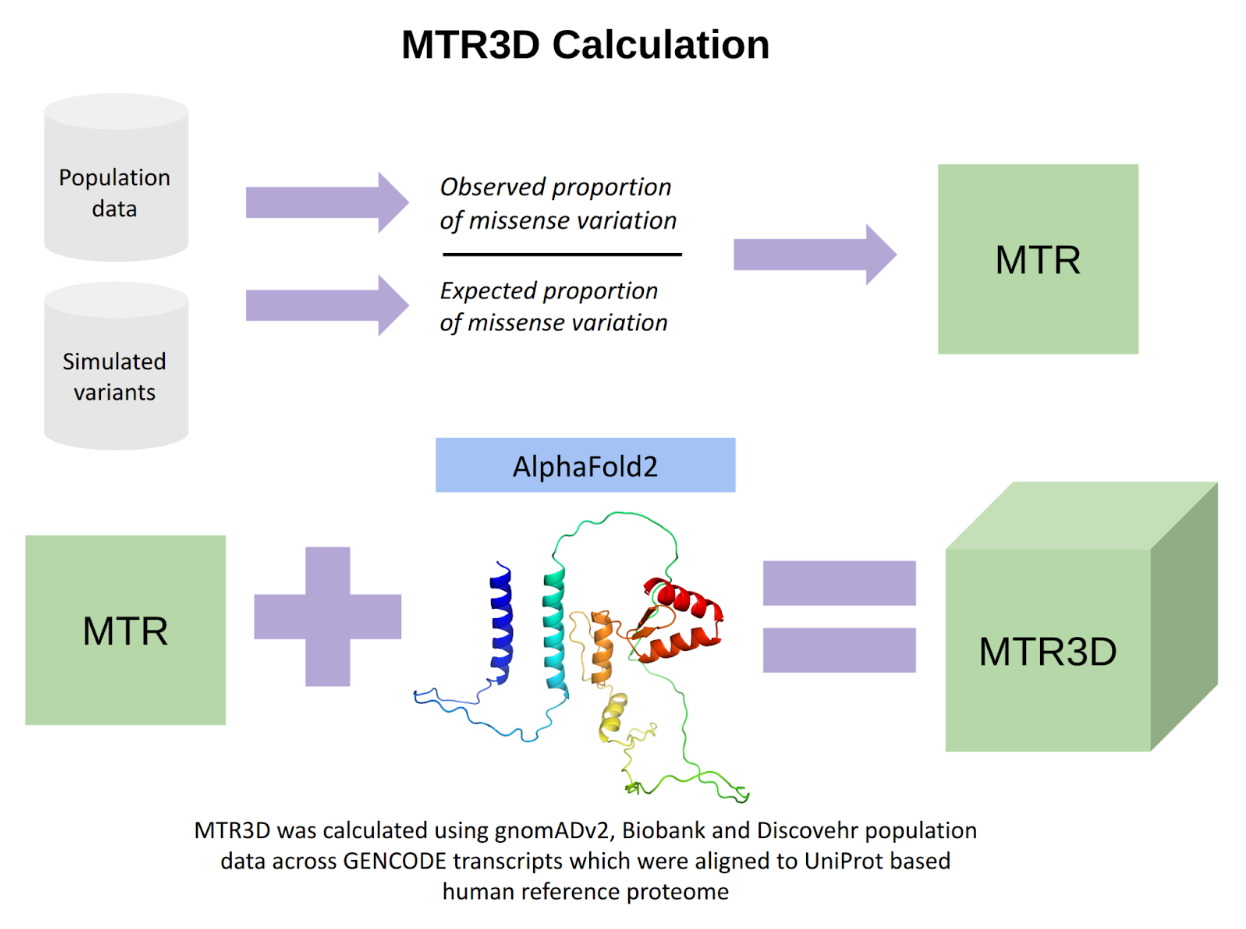
Abstract: The missense tolerance ratio (MTR) was developed as a novel approach to assess the deleteriousness of variants. Its three-dimensional successor, MTR3D, was demonstrated powerful at discriminating pathogenic from benign variants. However, its reliance on experimental structures and homologs limited its coverage of the proteome. We have now utilized AlphaFold2 models to develop MTR3D-AF2, which covers 89.31% of proteins and 85.39% of residues across the human proteome. This work has improved MTR3D's ability to distinguish clinically established pathogenic from benign variants. MTR3D-AF2 is freely available as an interactive web server at https://biosig.lab.uq.edu.au/mtr3daf2/