mmCSM-PPI is a valuable resource for the study of how multiple mutations affect protein-protein binding affinity.
Running predictions
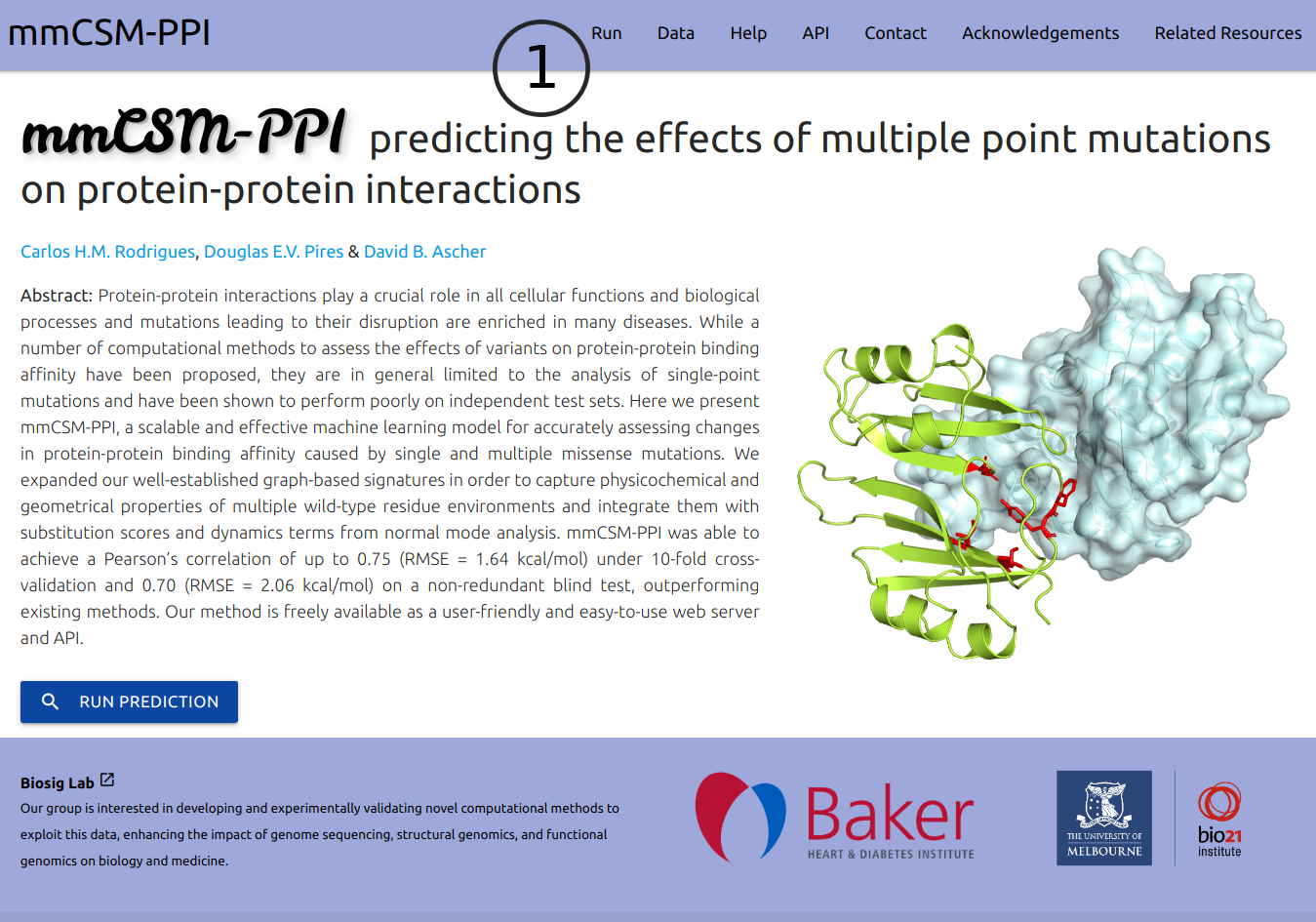
The input page can be accessed from the top menu Run (1).
Users are required to provide a protein structure by uploading a file in PDB format or typing a PDB four letter accession code (2).
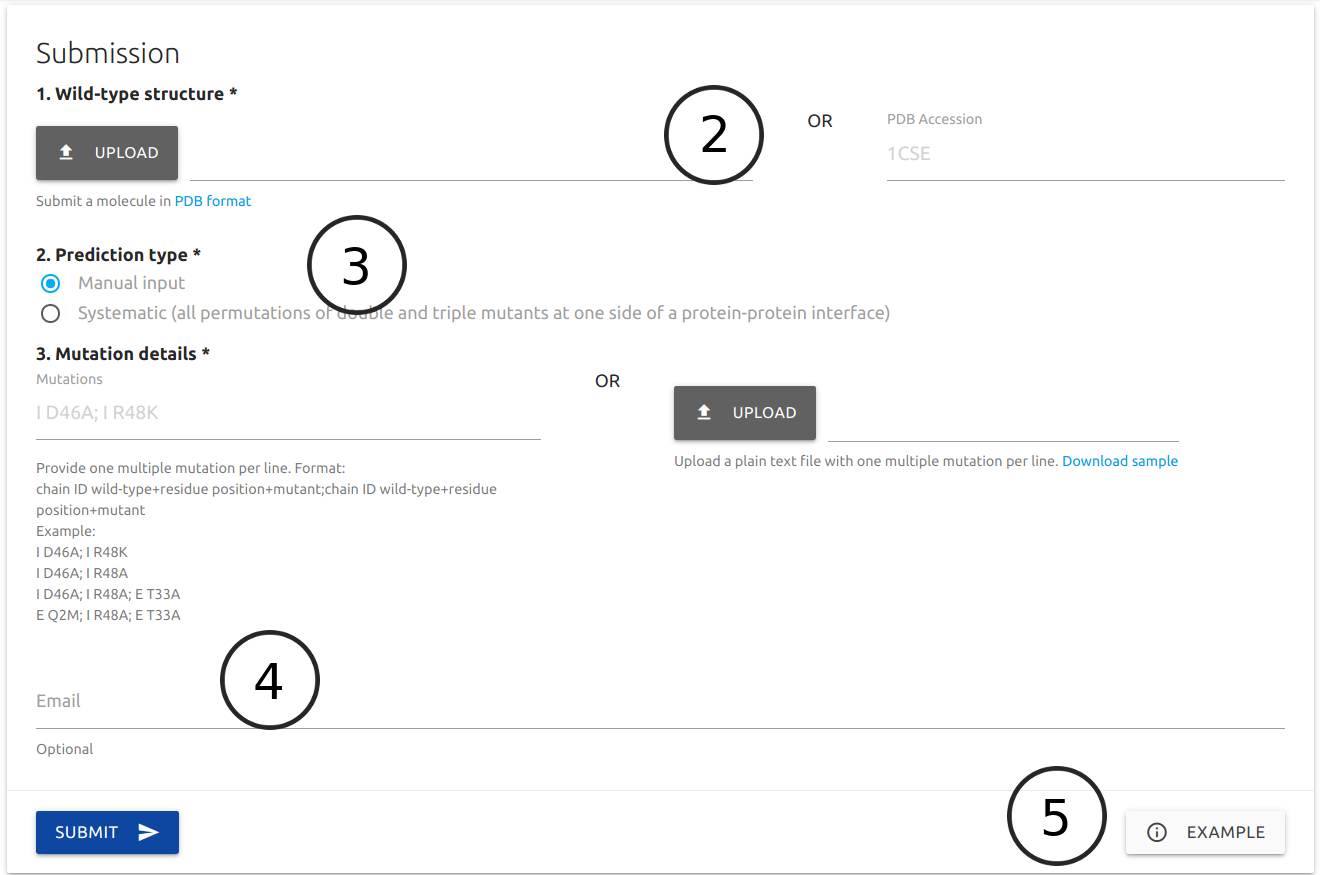
Two prediction types are available (3):
- Manual: This option allows for the input of specific multiple mutations which can be provided via the text field or by uploading a plain text file with a list of entries. In both cases, multiple mutations are must be separeted by a semi-colon (;) and one entry per line. Input examples and a sample file are available.
- Systematic: If this option is selected, users are required to provide a chain identifier from which interfaces residues will be identified and all permutations of double and triple mutants generated for evaluation. The results page will show top 100 entries increasing affinity and top 100 decreasing affinity.
If provided (4), an email will be sent to the user after the submission is processed.
A button for an example results page is available at the bottom of the form (5)
Results
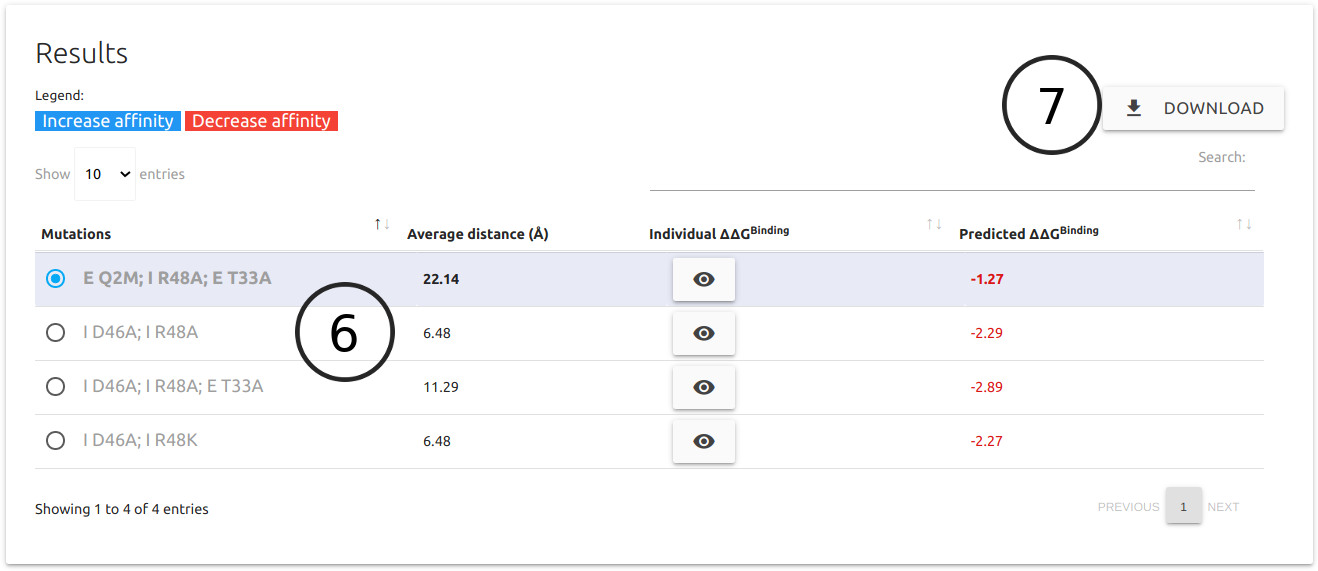
- For both types of prediction (manual and systematic), results are shown in table format (6) where entries predicted as increasing affinity are shown in blue and decreasing affinity in red. Individual ΔΔGBinding for each point mutation can also be viewed by clicking the button visibility. These are calculated using mCSM-PPI2.
- A button for downloading (7) all the results as a comma separated file (csv) is available
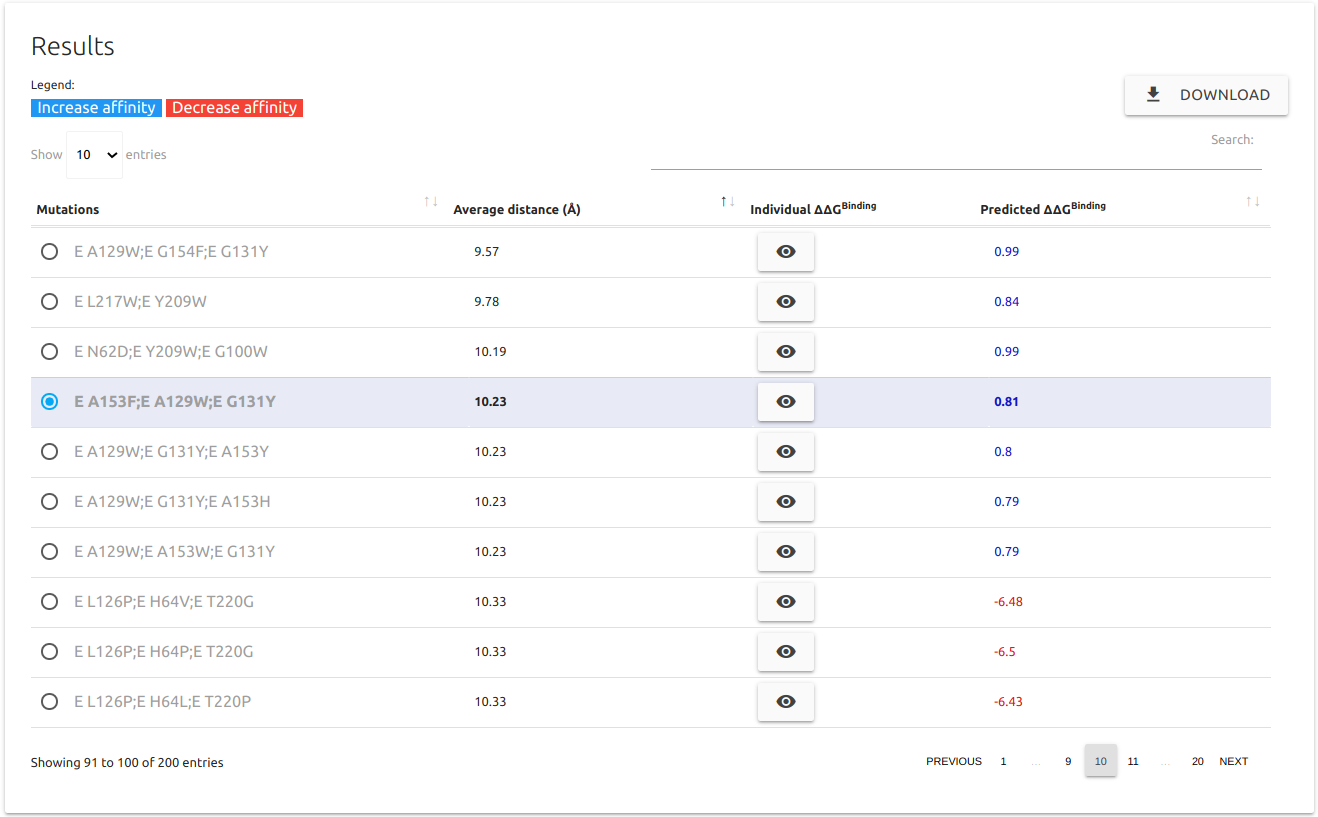
- Systematic evaluations of PPI interface shows the top 100 increasing/decreasing affinity entries based on all permutations of double and triple mutants at one side of a protein-protein interface.
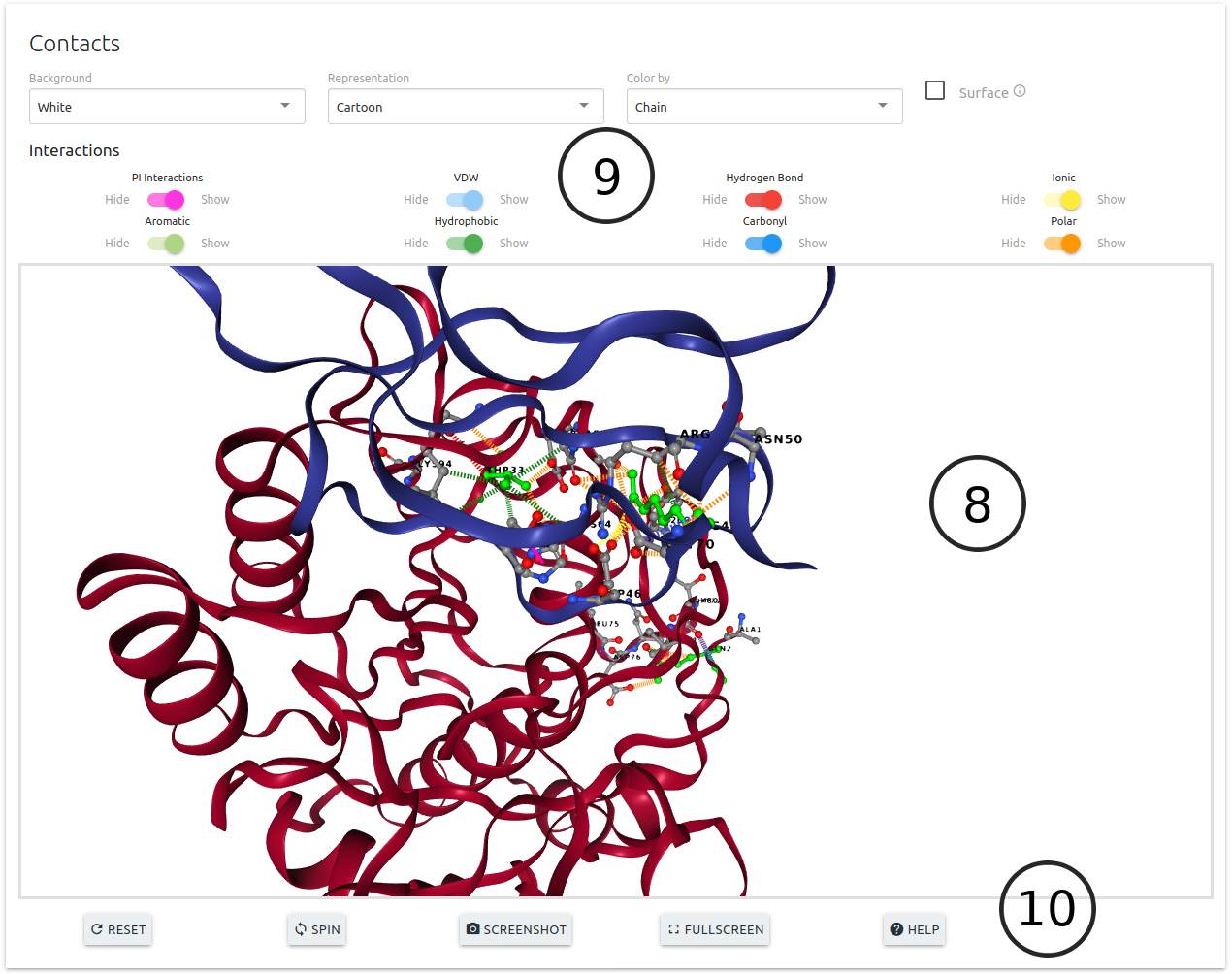
- An interactive 3D viewer is available (8) allowing for the analysis of interatomic interactions for the wild-type residues. A set of controls are also available for customising the viewer (9)
- Action buttons at the bottom of the viewer (10)
Contact us
In case you experience any troube using mmCSM-PPI or if you have any suggestions or comments, please do not hesitate in cantacting us either via email or via our Group website.
If your are contacting regarding a job submission, please include details such as input information and the job identifier