About mmCSM-Lig
Table of Contents
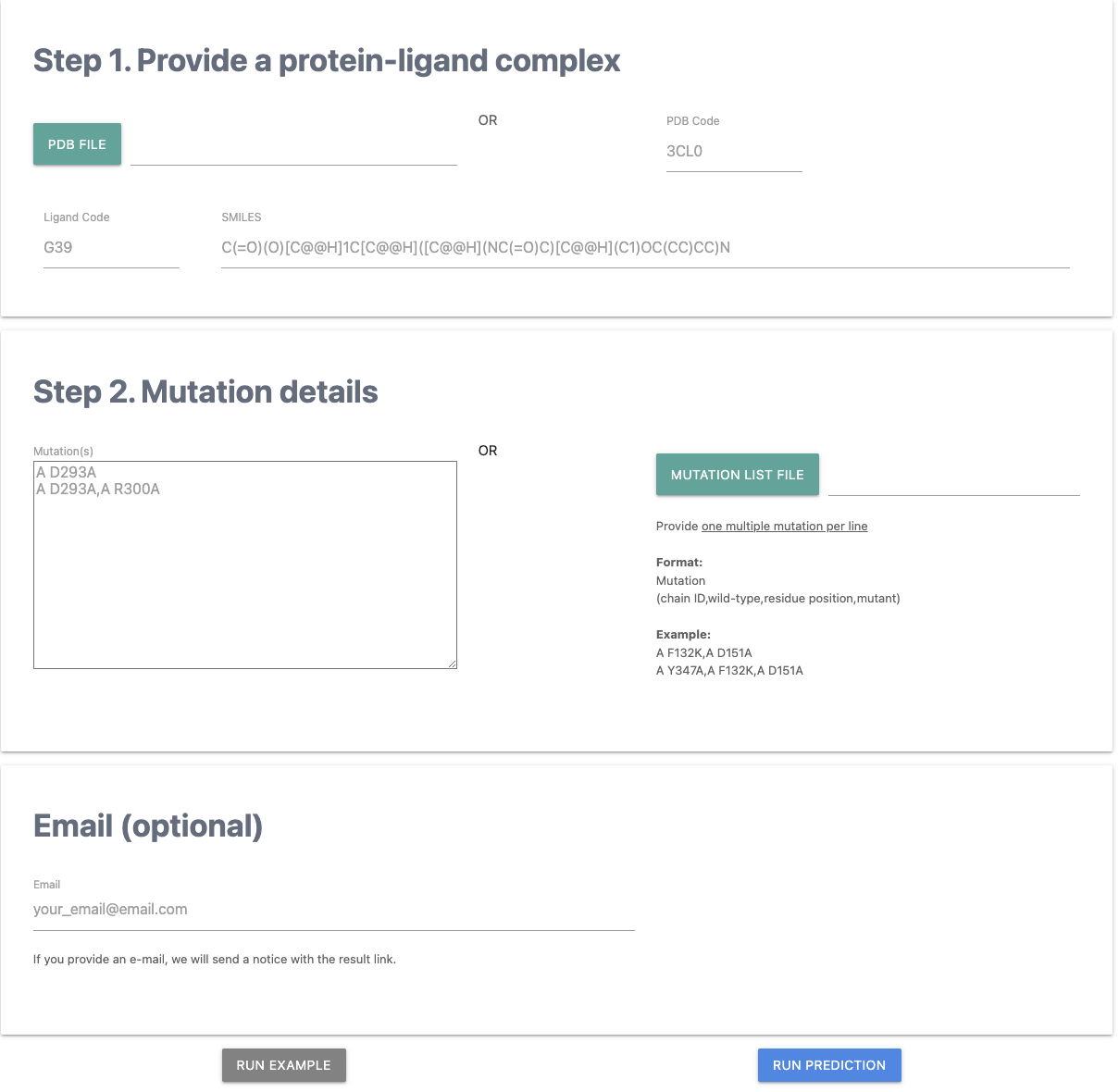
Submission
A. Users need to provide a protein-ligand 3D complex structure via either in a PDB file or a PDB accession. Also, the ligand information of the given complex structure such as Ligand Code (Three Letters) and SMILES needs to be provided.
B. At least one mutation has to be provided either via an "input tab" or a "file" with the format of "chain,wild-type,residue position,mutant". For more than one multiple mutation, it needs to be provided one multiple mutation per line.
C. Optionally, users can get a notice with the result link by providing an email.
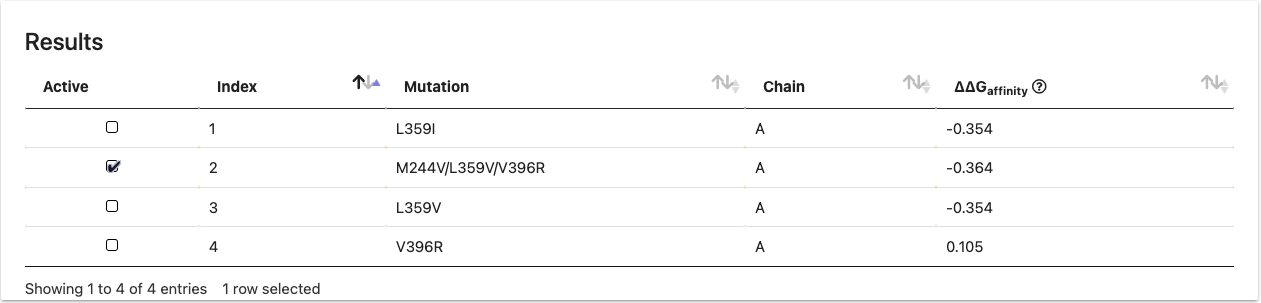
Results
Result Table
mmCSM-Lig enables users to browse the predicted ∆∆Gvalues and their 3D interaction views in a table and 3D molecule viewer.
A. The result table contains both mutational information and predicted values.
B. Users can visualise the molecular interactions of mutation sites of each mutation in 3D viewer by selecting in the table.
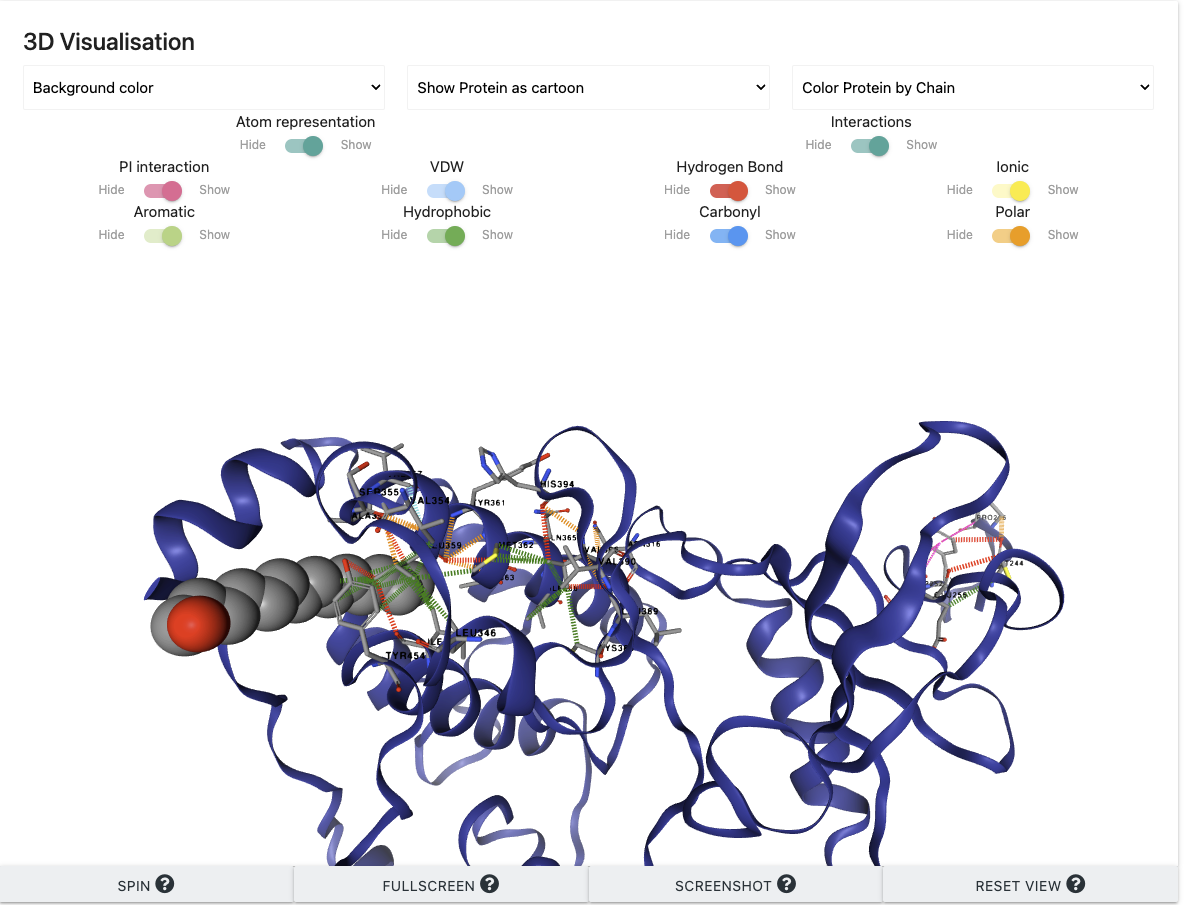
3D Interaction Viewer
The 3D viewer shows a protein-ligand complex with molecular interactions of mutation sites.
A. Protein can be shown in different representations and colours in the drop-down menu.
B. Users can control the visualisation of moleculear interactions and the residues participating in the interactions with buttons.
Download Results
The result table and 3D visualisation are downloable in a CSV and PSE format, respectively.
