Help - How to use mCSM-AB
Main page
About mCSM-AB
mCSM-AB is a novel computational method to predict the change in antibody-antigen affinity (in terms of a ∆∆G) upon the introduction
of a single mutation. It is a machine learning approach that relies on graph-based structural signatures.
Such a predictive model is not only of great relevance for antibody engineering and development, but would also allow
the prediction of biologically relevant escape mutations.
Submission page
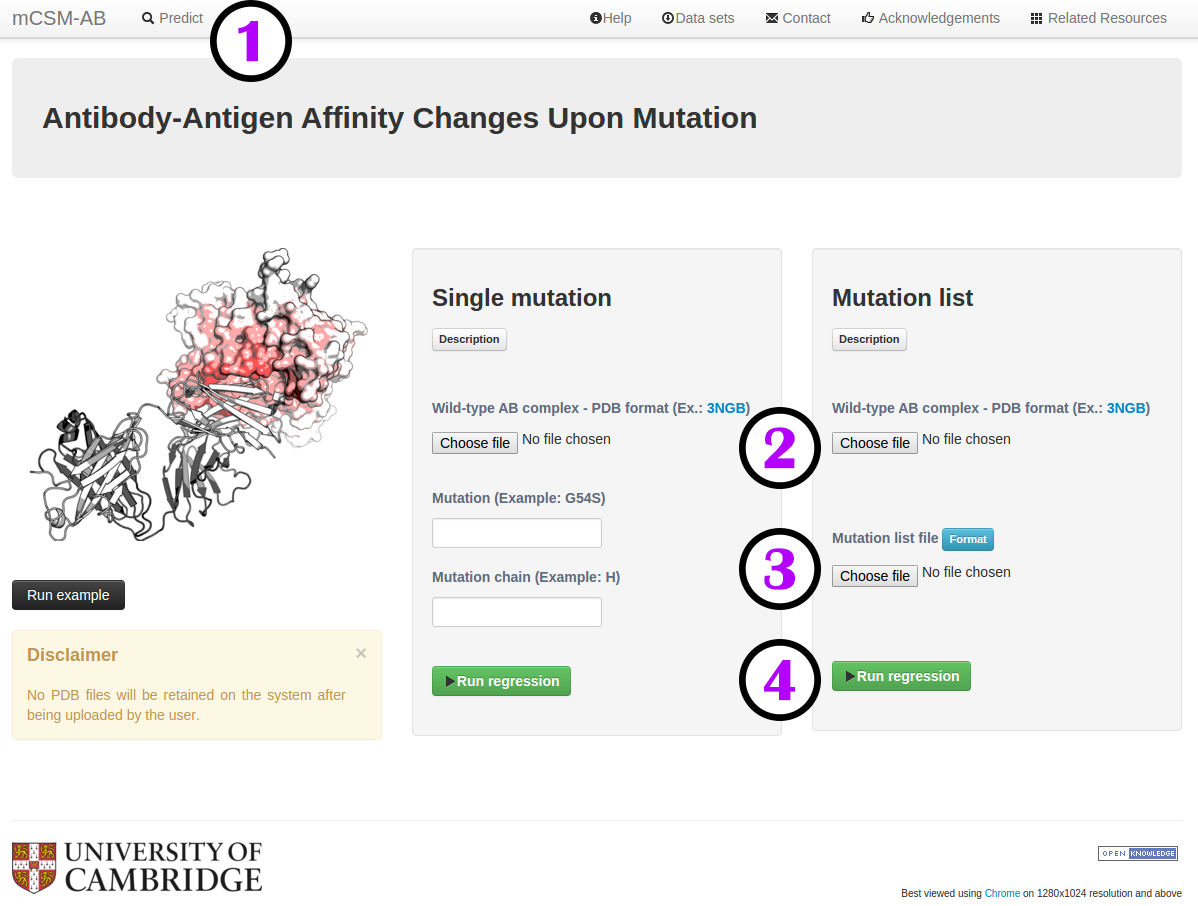
How to run a prediction
To run a prediction:
- Click on "Predict" (1) to open the submission page.
- Provide the structure of the wild-type antibody-antigen complex (2), which must comply with the PDB format.
- A single mutation or a file with a list of mutations to be analysed should be provided (3). For single mutation mode, a mutation code consists of wild-type code, residue position and mutant code (using the one letter amino acid code). Residue position must be consistent with the PDB file. The chain for the mutation code must also be provided.
- You are then ready to submit your query for analysis (4).
Results page
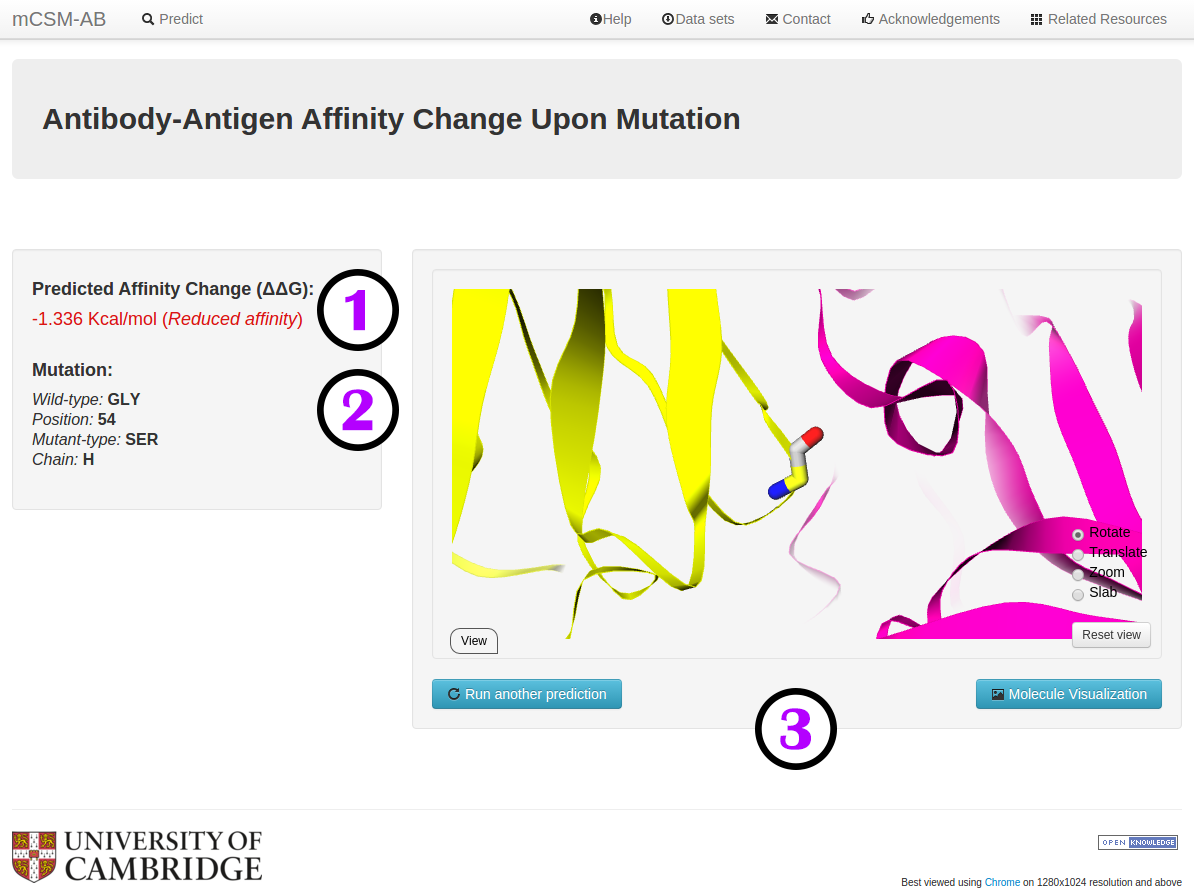
Results - Single mutation
Your results (1) for a single mutation will be displayed once computations are completed. The results will display the predicted
change in affinity upon mutation (ΔΔG in Kcal/mol). A negative value (and red writing) corresponds
to a mutation predicted as destabilising; while a positive sign (and blue writing) corresponds to a mutation
predicted as stabilising. Complementary information also displayed include:
- A summary of the mutation is presented (2) highlighting the wild-type residue, position number and chain.
- The protein complex and mutation can be visualised directly from the server (3).
Results page
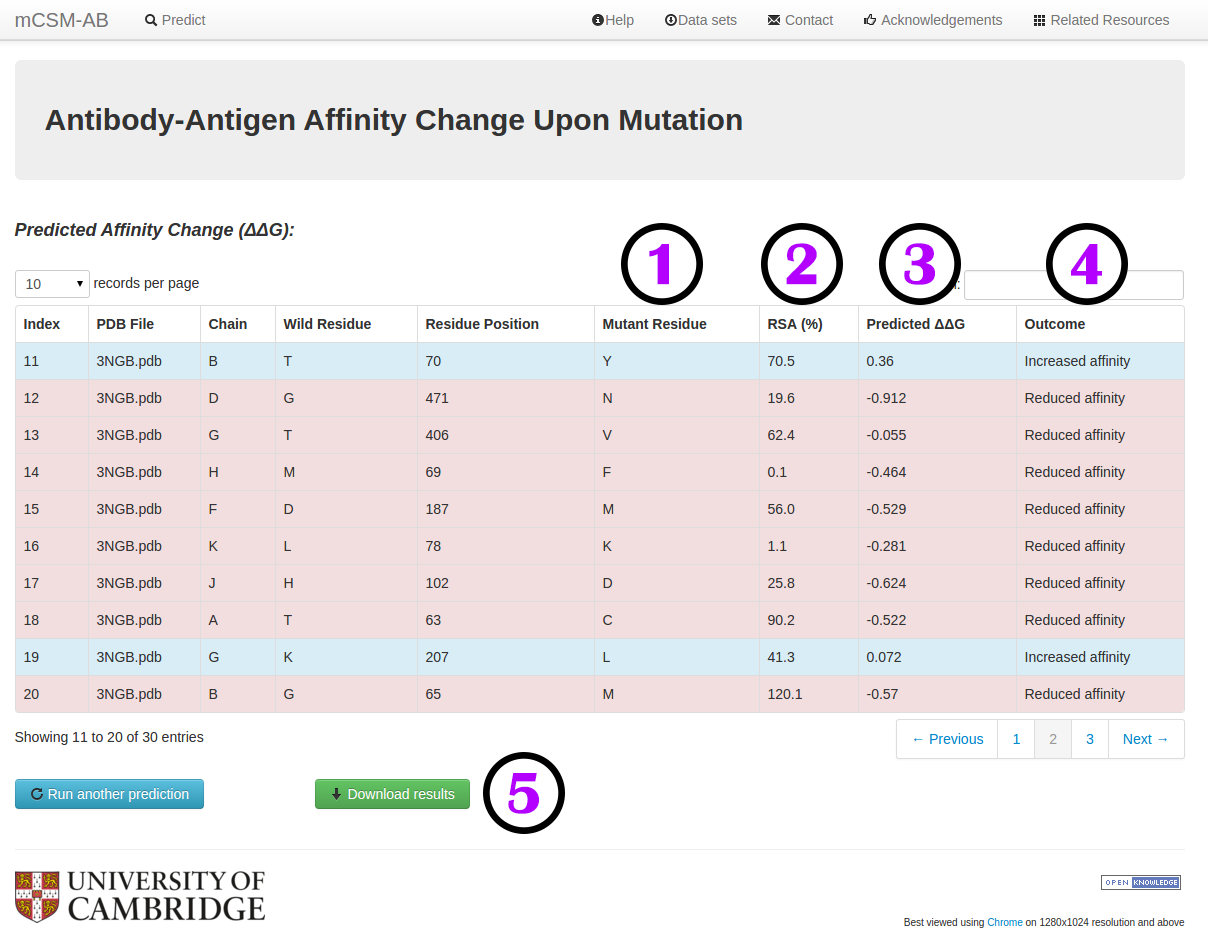
Results - List of Mutations
Your results for a list of mutations will be displayed in a table format with the following information:
- Mutant residue code (1).
- Residue relative solvent accessibility (RSA) (2).
- The predicted ΔΔG (3) and the predicted outcome (4).
Contact page
Getting in touch
In case you experience any trouble using mCSM-AB or have any suggestions or comments, please do not hesitate in contacting us (1) either via e-mail or through the online form.