CSM-Potential a deep learning platform for identification of PrOTEin iNteracTIons And biological Ligands using 3D structures
Carlos H.M. Rodrigues & David B. Ascher
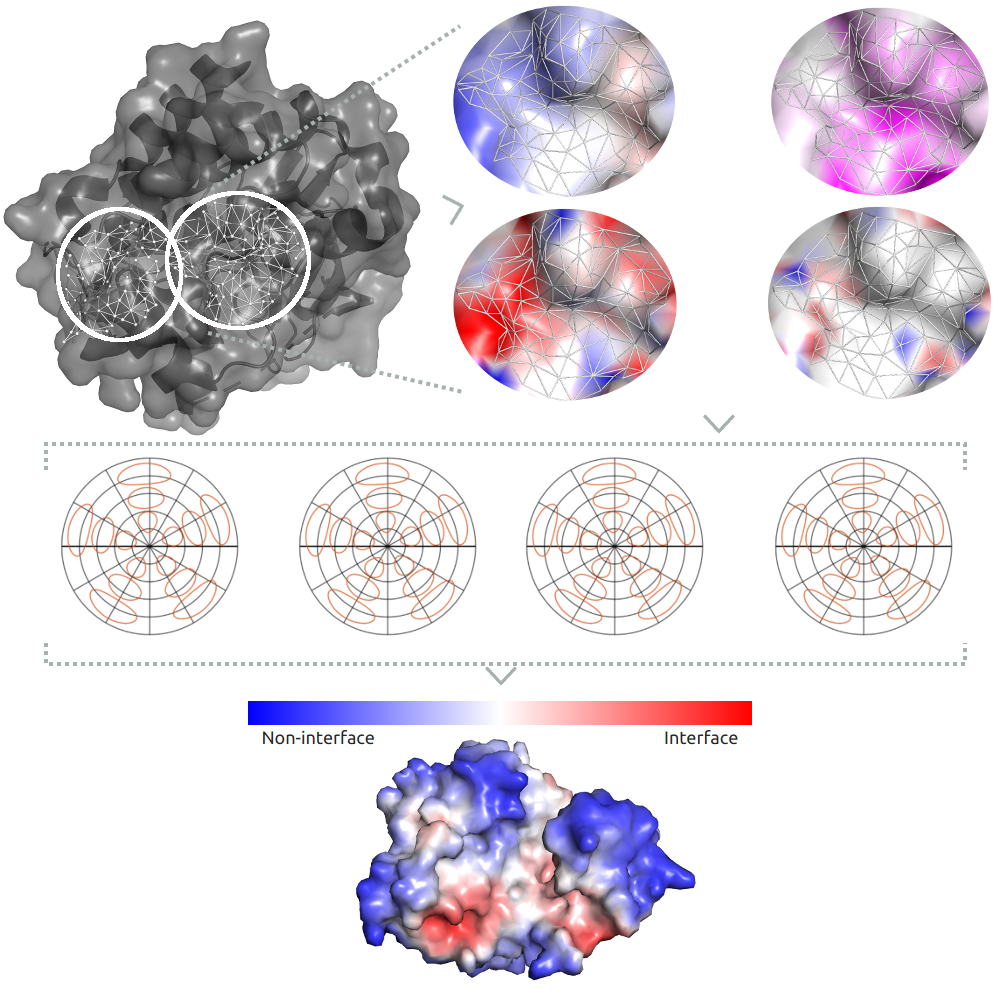
Abstract: Recent advances in protein structural modelling have enabled the accurate prediction of the holo 3D structures of almost any protein, however protein function is intrinsically linked to the interactions it makes. While a number of computational approaches have been proposed to explore potential biological interactions, they have been limited to specific interactions, and have not been readily accessible for non-experts or use in bioinformatic pipelines. Here we present CSM-Potential, a geometric deep learning approach to identify regions of a protein surface that are likely to mediate protein-protein and protein-ligand interactions in order to provide a link between 3D structure and biological function. Our method has shown robust performance, outperforming existing methods for both predictive tasks. By assessing the performance of CSM-Potential on independent blind tests, we show that our method was able to achieve ROC AUC values up to 0.81 the identification of potential protein-protein binding sites and up to 0.96 accuracy on biological ligand classification.
This website is free for all users. For  policy please click here.
policy please click here.