Help - How to use CSM-lig
Main page
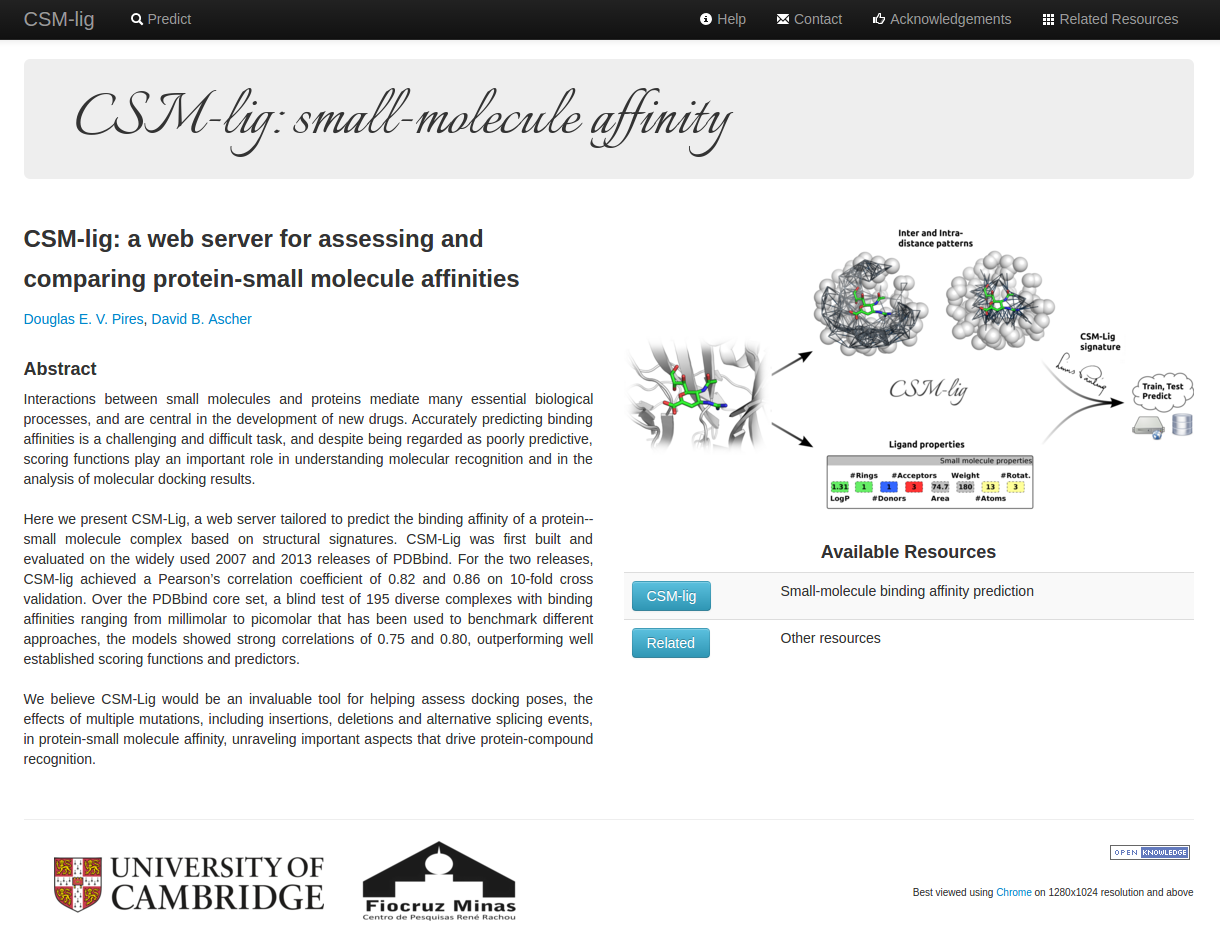
About CSM-lig
CSM-lig is a machine-learning method to predict affinities of small-molecules to proteins, which relies
on distance/pharmacophore patterns encoded as graph-based signatures.
A user-friendly and freely accessible web interface was developed and allows users to submit either a single
PDB structure of the protein-small molecule complex, or a compressed file with multiple structures, which could
include different poses of the same complex, different ligands or multiple different proteins.
Submission page
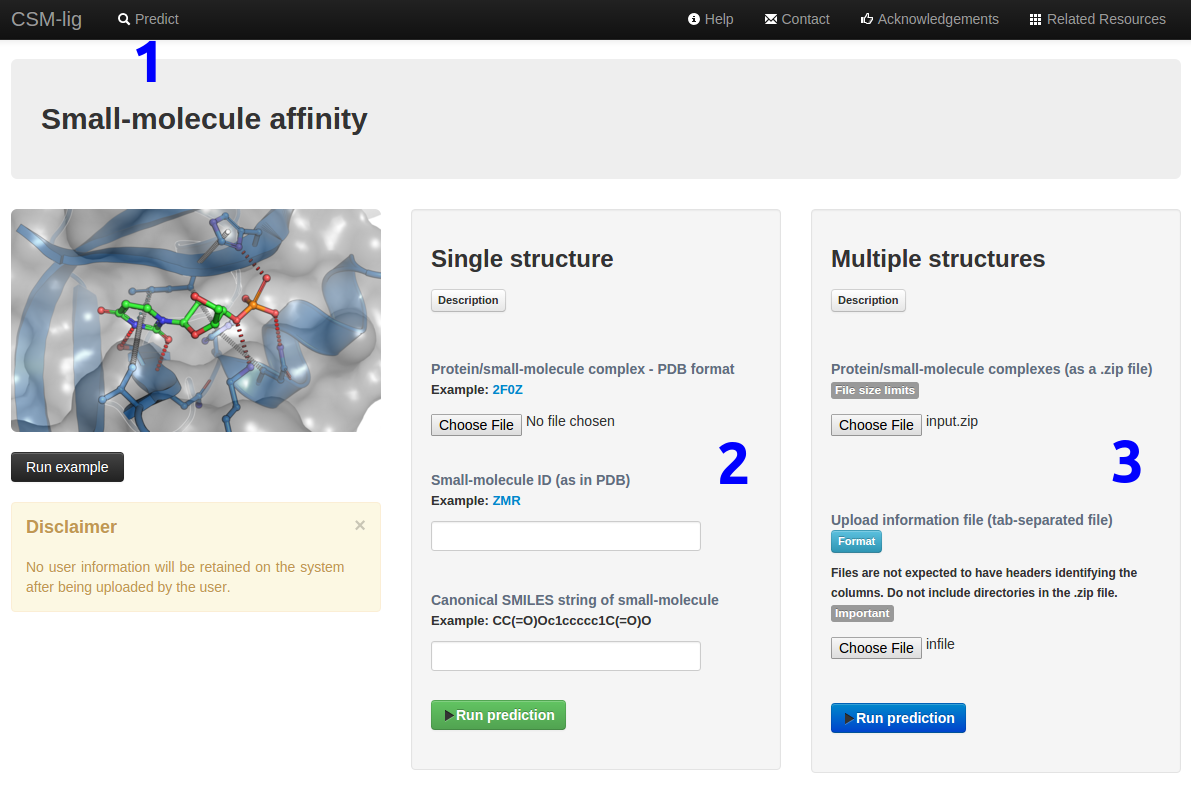
How to run a prediction
To run a prediction:
- Click on "Predict" (1) to open the submission page.
- Provide either:
- A single PDB file and the small-molecule information (HET ID and SMILES string) (2) or
- As input files, a compressed file with PDBs (.zip) and a tab-separated file with the molecule information (HET ID and SMILES string for each uploaded PDB)(3).
Parsing Considerations
* Water molecules present in the uploaded PDB files are removed prior to calculation. * Cofactors, metals and ions are also removed from the structure and are not considered in the structural signature calculations.Results page
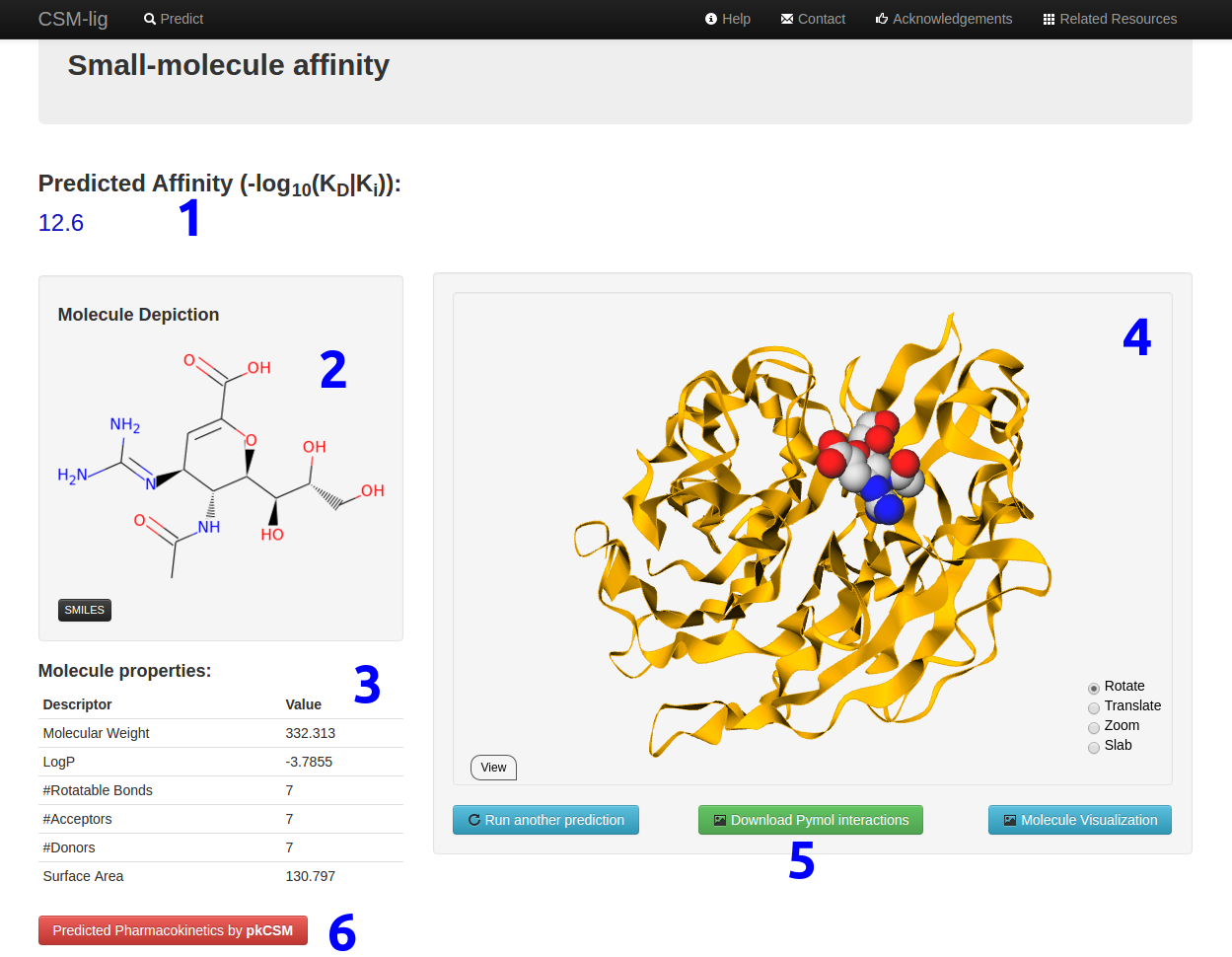
Results
For a single molecule prediction, your results can be displayed as follows:
- The affinity prediction will be shown in (1).
- A depiction of the small-molecule will be shown in (2). Make sure the depiction is what you are expecting for your molecule.
- A list of molecule properties will be calculated and shown in (3).
- An interactive molecular visualization of the uploaded complex will be shown in (4).
- Users have the option to download a Pymol session file for visualizing the ligand interactions in (5).
- There is also an option of calculating the phamacokinetic and toxicity properties of the ligand using the pkCSM server (6).
Contact page
Getting in touch
In case you experience any trouble using CSM-lig or have any suggestions or comments, please do not hesitate in contacting us either via e-mail or through the online form.