Non-structural protein 7 (nsp7) (Uniprot: P0DTD1 - GenBank: QHD43415_7)
MTR: 0.76 (Intolerant) / RVIS: -0.12 (Intolerant)
Sequence
Mutational Tolerance (Missense Tolerance Ratio)
Horizontal lines show gene-specific MTR percentiles 5th, 25th, 50th, and neutrality (MTR = 1.0)
Mutations launchAnalysis
launchSaturation Mutagenesis
info_outline
SARS-CoV2 Circulating Variants
| Position | AA from | AA to | Chain | Frequency (%) | Depth (Å) | Phi (°) | Psi (°) | RSA (%) | SST | MTR Score | Consurf Score | ΔAtomic Fluctuation (Å) | ΔEnergy Deformation (kcal/mol) | ΔΔGmCSMstability | ΔΔGDynaMutstability | ΔΔGDUETstability | ΔΔGSDMstability | ΔΔGENCoMstability | ΔΔSENCoMvib | Hydrophobics | Positives | Negatives | Acceptors | Donnors | Aromatics | Sulfurs | Neutral | ΔHydrogen bond | ΔIonic | ΔAromatic | ΔHydrophobic | ΔCarbonyl | ΔPolar | ΔPi | blosum100 | blosum30 | blosum35 | blosum40 | blosum45 | blosum50 | blosum55 | blosum60 | blosum62 | blosum65 | blosum70 | blosum75 | blosum80 | blosum85 | blosum90 | blosum95 | pam120 | pam180 | pam250 | pam30 | pam300 | pam60 | pam90 | HENS920102 | HENS920103 | WEIL970102 | HENS920101 | GRAR740104 | PRLA000102 | HENS920104 | WEIL970101 | GIAG010101 | RISJ880101 | DOSZ010102 | DOSZ010103 | JOND940101 | DOSZ010101 | ZHAC000101 | MIYS990106 | ZHAC000103 | LIWA970101 | QU_C930103 | QU_C930102 | QU_C930101 | ZHAC000106 | ZHAC000104 | MIYS850102 | MIYS850103 | MIYS990107 | GODA950101 | BRYS930101 | RIER950101 | KOSJ950110_RSA | JOND920103 | VOGG950101 | KESO980102 | OGAK980101 | NIEK910102 | AZAE970101 | AZAE970102 | NIEK910101 | TUDE900101 | CROG050101 | MIYS930101 | MIYS960101 | SIMK990105 | SIMK990104 | TOBD000102 | BETM990101 | TOBD000101 | KOLA920101 | MEHP950102 | MEHP950101 | PRLA000101 | JOHM930101 | QUIB020101 | ROBB790102 | MIRL960101 | GEOD900101 | KOSJ950100_SST | DAYM780302 | DAYM780301 | LUTR910106 | LUTR910107 | LUTR910104 | LUTR910105 | LUTR910102 | LUTR910103 | THOP960101 | LUTR910101 | GONG920101 | MCLA710101 | LUTR910108 | LUTR910109 | SKOJ970101 | SKOJ000101 | OVEJ920101 | KANM000101 | VENM980101 | RUSR970103 | RUSR970102 | RUSR970101 | LINK010101 | MUET010101 | KOLA930101 | MCLA720101 | ALTS910101 | LEVJ860101 | BONM030101 | BONM030106 | MOOG990101 | BONM030105 | ZHAC000105 | KAPO950101 | OVEJ920100_RSA | FEND850101 | NGPC000101 | SIMK990103 | KESO980101 | SKOJ000102 | TANS760101 | TANS760102 | MIYT790101 | BLAJ010101 | PARB960101 | CSEM940101 | OVEJ920102 | OVEJ920103 | BONM030103 | PARB960102 | MICC010101 | BONM030102 | KOSJ950100_RSA_SST | DOSZ010104 | MUET020101 | MUET020102 | ZHAC000102 | SIMK990102 | NAOD960101 | SIMK990101 | MEHP950103 | MIYS960102 | MIYS960103 | BONM030104 | KOSJ950115 | BASU010101 | BENS940104 | BENS940102 | BENS940103 | BENS940101 | FITW660101 | MOHR870101 | A | A.1 | A.1.1 | A.1.3 | A.2 | A.3 | A.4 | A.5 | A.6 | B | B.1 | B.1.1 | B.1.1.1 | B.1.1.10 | B.1.1.13 | B.1.1.14 | B.1.1.17 | B.1.1.18 | B.1.1.2 | B.1.1.3 | B.1.1.4 | B.1.1.5 | B.1.1.6 | B.1.1.7 | B.1.1.8 | B.1.1.9 | B.1.12 | B.1.13 | B.1.19 | B.1.22 | B.1.23 | B.1.26 | B.1.29 | B.1.3 | B.1.30 | B.1.31 | B.1.32 | B.1.33 | B.1.34 | B.1.35 | B.1.36 | B.1.37 | B.1.38 | B.1.39 | B.1.40 | B.1.41 | B.1.43 | B.1.44 | B.1.5 | B.1.5.1 | B.1.5.2 | B.1.5.3 | B.1.5.4 | B.1.5.5 | B.1.5.6 | B.1.6 | B.1.66 | B.1.67 | B.1.69 | B.1.70 | B.1.71 | B.1.72 | B.1.8 | B.10 | B.13 | B.14 | B.15 | B.16 | B.2 | B.2.1 | B.2.2 | B.2.4 | B.2.5 | B.2.6 | B.2.7 | B.3 | B.4 | B.5 | B.6 | B.7 | B.9 | Details |
|---|
Mutations from SARS info_outline
| Position | AA from | AA to | Chain | Frequency (%) | Depth (Å) | Phi (°) | Psi (°) | RSA (%) | SST | MTR Score | Consurf Score | ΔAtomic Fluctuation (Å) | ΔEnergy Deformation (kcal/mol) | ΔΔGmCSMstability | ΔΔGDynaMutstability | ΔΔGDUETstability | ΔΔGSDMstability | ΔΔGENCoMstability | ΔΔSENCoMvib | Hydrophobics | Positives | Negatives | Acceptors | Donnors | Aromatics | Sulfurs | Neutral | Hydrogen bond | Ionic | Aromatic | Hydrophobic | Carbonyl | Polar | Pi | blosum100 | blosum30 | blosum35 | blosum40 | blosum45 | blosum50 | blosum55 | blosum60 | blosum62 | blosum65 | blosum70 | blosum75 | blosum80 | blosum85 | blosum90 | blosum95 | pam120 | pam180 | pam250 | pam30 | pam300 | pam60 | pam90 | HENS920102 | HENS920103 | WEIL970102 | HENS920101 | GRAR740104 | PRLA000102 | HENS920104 | WEIL970101 | GIAG010101 | RISJ880101 | DOSZ010102 | DOSZ010103 | JOND940101 | DOSZ010101 | ZHAC000101 | MIYS990106 | ZHAC000103 | LIWA970101 | QU_C930103 | QU_C930102 | QU_C930101 | ZHAC000106 | ZHAC000104 | MIYS850102 | MIYS850103 | MIYS990107 | GODA950101 | BRYS930101 | RIER950101 | KOSJ950110_RSA | JOND920103 | VOGG950101 | KESO980102 | OGAK980101 | NIEK910102 | AZAE970101 | AZAE970102 | NIEK910101 | TUDE900101 | CROG050101 | MIYS930101 | MIYS960101 | SIMK990105 | SIMK990104 | TOBD000102 | BETM990101 | TOBD000101 | KOLA920101 | MEHP950102 | MEHP950101 | PRLA000101 | JOHM930101 | QUIB020101 | ROBB790102 | MIRL960101 | GEOD900101 | KOSJ950100_SST | DAYM780302 | DAYM780301 | LUTR910106 | LUTR910107 | LUTR910104 | LUTR910105 | LUTR910102 | LUTR910103 | THOP960101 | LUTR910101 | GONG920101 | MCLA710101 | LUTR910108 | LUTR910109 | SKOJ970101 | SKOJ000101 | OVEJ920101 | KANM000101 | VENM980101 | RUSR970103 | RUSR970102 | RUSR970101 | LINK010101 | MUET010101 | KOLA930101 | MCLA720101 | ALTS910101 | LEVJ860101 | BONM030101 | BONM030106 | MOOG990101 | BONM030105 | ZHAC000105 | KAPO950101 | OVEJ920100_RSA | FEND850101 | NGPC000101 | SIMK990103 | KESO980101 | SKOJ000102 | TANS760101 | TANS760102 | MIYT790101 | BLAJ010101 | PARB960101 | CSEM940101 | OVEJ920102 | OVEJ920103 | BONM030103 | PARB960102 | MICC010101 | BONM030102 | KOSJ950100_RSA_SST | DOSZ010104 | MUET020101 | MUET020102 | ZHAC000102 | SIMK990102 | NAOD960101 | SIMK990101 | MEHP950103 | MIYS960102 | MIYS960103 | BONM030104 | KOSJ950115 | BASU010101 | BENS940104 | BENS940102 | BENS940103 | BENS940101 | FITW660101 | MOHR870101 | A | A.1 | A.1.1 | A.1.3 | A.2 | A.3 | A.4 | A.5 | A.6 | B | B.1 | B.1.1 | B.1.1.1 | B.1.1.10 | B.1.1.13 | B.1.1.14 | B.1.1.17 | B.1.1.18 | B.1.1.2 | B.1.1.3 | B.1.1.4 | B.1.1.5 | B.1.1.6 | B.1.1.7 | B.1.1.8 | B.1.1.9 | B.1.12 | B.1.13 | B.1.19 | B.1.22 | B.1.23 | B.1.26 | B.1.29 | B.1.3 | B.1.30 | B.1.31 | B.1.32 | B.1.33 | B.1.34 | B.1.35 | B.1.36 | B.1.37 | B.1.38 | B.1.39 | B.1.40 | B.1.41 | B.1.43 | B.1.44 | B.1.5 | B.1.5.1 | B.1.5.2 | B.1.5.3 | B.1.5.4 | B.1.5.5 | B.1.5.6 | B.1.6 | B.1.66 | B.1.67 | B.1.69 | B.1.70 | B.1.71 | B.1.72 | B.1.8 | B.10 | B.13 | B.14 | B.15 | B.16 | B.2 | B.2.1 | B.2.2 | B.2.4 | B.2.5 | B.2.6 | B.2.7 | B.3 | B.4 | B.5 | B.6 | B.7 | B.9 | Details |
|---|
Mutations from Bat RaTG13 info_outline
| Position | AA from | AA to | Chain | Frequency (%) | Depth (Å) | Phi (°) | Psi (°) | RSA (%) | SST | MTR Score | Consurf Score | ΔAtomic Fluctuation (Å) | ΔEnergy Deformation (kcal/mol) | ΔΔGmCSMstability | ΔΔGDynaMutstability | ΔΔGDUETstability | ΔΔGSDMstability | ΔΔGENCoMstability | ΔΔSENCoMvib | Hydrophobics | Positives | Negatives | Acceptors | Donnors | Aromatics | Sulfurs | Neutral | Hydrogen bond | Ionic | Aromatic | Hydrophobic | Carbonyl | Polar | Pi | blosum100 | blosum30 | blosum35 | blosum40 | blosum45 | blosum50 | blosum55 | blosum60 | blosum62 | blosum65 | blosum70 | blosum75 | blosum80 | blosum85 | blosum90 | blosum95 | pam120 | pam180 | pam250 | pam30 | pam300 | pam60 | pam90 | HENS920102 | HENS920103 | WEIL970102 | HENS920101 | GRAR740104 | PRLA000102 | HENS920104 | WEIL970101 | GIAG010101 | RISJ880101 | DOSZ010102 | DOSZ010103 | JOND940101 | DOSZ010101 | ZHAC000101 | MIYS990106 | ZHAC000103 | LIWA970101 | QU_C930103 | QU_C930102 | QU_C930101 | ZHAC000106 | ZHAC000104 | MIYS850102 | MIYS850103 | MIYS990107 | GODA950101 | BRYS930101 | RIER950101 | KOSJ950110_RSA | JOND920103 | VOGG950101 | KESO980102 | OGAK980101 | NIEK910102 | AZAE970101 | AZAE970102 | NIEK910101 | TUDE900101 | CROG050101 | MIYS930101 | MIYS960101 | SIMK990105 | SIMK990104 | TOBD000102 | BETM990101 | TOBD000101 | KOLA920101 | MEHP950102 | MEHP950101 | PRLA000101 | JOHM930101 | QUIB020101 | ROBB790102 | MIRL960101 | GEOD900101 | KOSJ950100_SST | DAYM780302 | DAYM780301 | LUTR910106 | LUTR910107 | LUTR910104 | LUTR910105 | LUTR910102 | LUTR910103 | THOP960101 | LUTR910101 | GONG920101 | MCLA710101 | LUTR910108 | LUTR910109 | SKOJ970101 | SKOJ000101 | OVEJ920101 | KANM000101 | VENM980101 | RUSR970103 | RUSR970102 | RUSR970101 | LINK010101 | MUET010101 | KOLA930101 | MCLA720101 | ALTS910101 | LEVJ860101 | BONM030101 | BONM030106 | MOOG990101 | BONM030105 | ZHAC000105 | KAPO950101 | OVEJ920100_RSA | FEND850101 | NGPC000101 | SIMK990103 | KESO980101 | SKOJ000102 | TANS760101 | TANS760102 | MIYT790101 | BLAJ010101 | PARB960101 | CSEM940101 | OVEJ920102 | OVEJ920103 | BONM030103 | PARB960102 | MICC010101 | BONM030102 | KOSJ950100_RSA_SST | DOSZ010104 | MUET020101 | MUET020102 | ZHAC000102 | SIMK990102 | NAOD960101 | SIMK990101 | MEHP950103 | MIYS960102 | MIYS960103 | BONM030104 | KOSJ950115 | BASU010101 | BENS940104 | BENS940102 | BENS940103 | BENS940101 | FITW660101 | MOHR870101 | A | A.1 | A.1.1 | A.1.3 | A.2 | A.3 | A.4 | A.5 | A.6 | B | B.1 | B.1.1 | B.1.1.1 | B.1.1.10 | B.1.1.13 | B.1.1.14 | B.1.1.17 | B.1.1.18 | B.1.1.2 | B.1.1.3 | B.1.1.4 | B.1.1.5 | B.1.1.6 | B.1.1.7 | B.1.1.8 | B.1.1.9 | B.1.12 | B.1.13 | B.1.19 | B.1.22 | B.1.23 | B.1.26 | B.1.29 | B.1.3 | B.1.30 | B.1.31 | B.1.32 | B.1.33 | B.1.34 | B.1.35 | B.1.36 | B.1.37 | B.1.38 | B.1.39 | B.1.40 | B.1.41 | B.1.43 | B.1.44 | B.1.5 | B.1.5.1 | B.1.5.2 | B.1.5.3 | B.1.5.4 | B.1.5.5 | B.1.5.6 | B.1.6 | B.1.66 | B.1.67 | B.1.69 | B.1.70 | B.1.71 | B.1.72 | B.1.8 | B.10 | B.13 | B.14 | B.15 | B.16 | B.2 | B.2.1 | B.2.2 | B.2.4 | B.2.5 | B.2.6 | B.2.7 | B.3 | B.4 | B.5 | B.6 | B.7 | B.9 | Details |
|---|
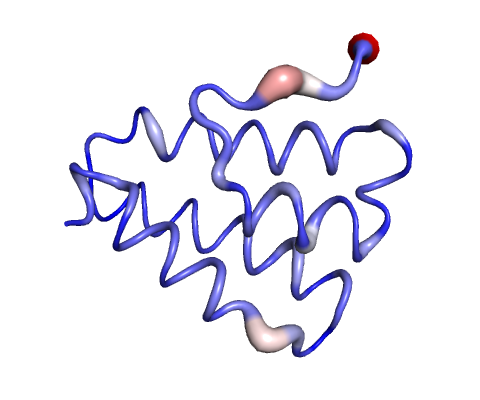
Molecular Dynamics
Downloads info_outline