Learn more about ThermoMutDB in less than 3 minutes.
Check some examples of using ThermoMutDB API
You can use the API form to choose an endpoint and setup parameters to get data, like in the following examples:
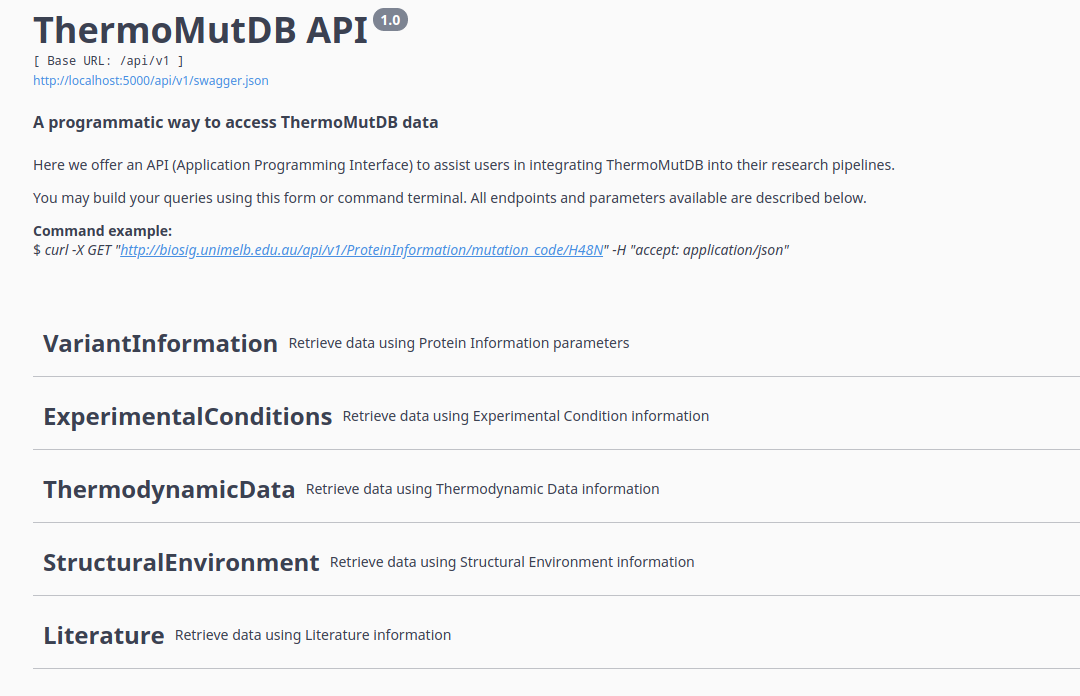
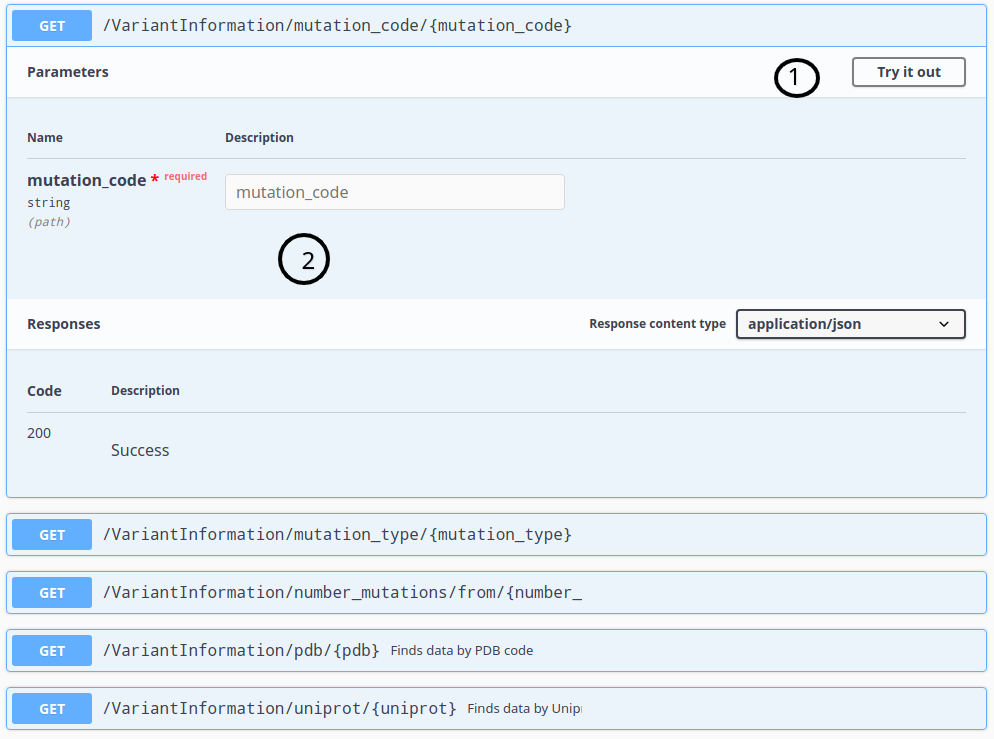
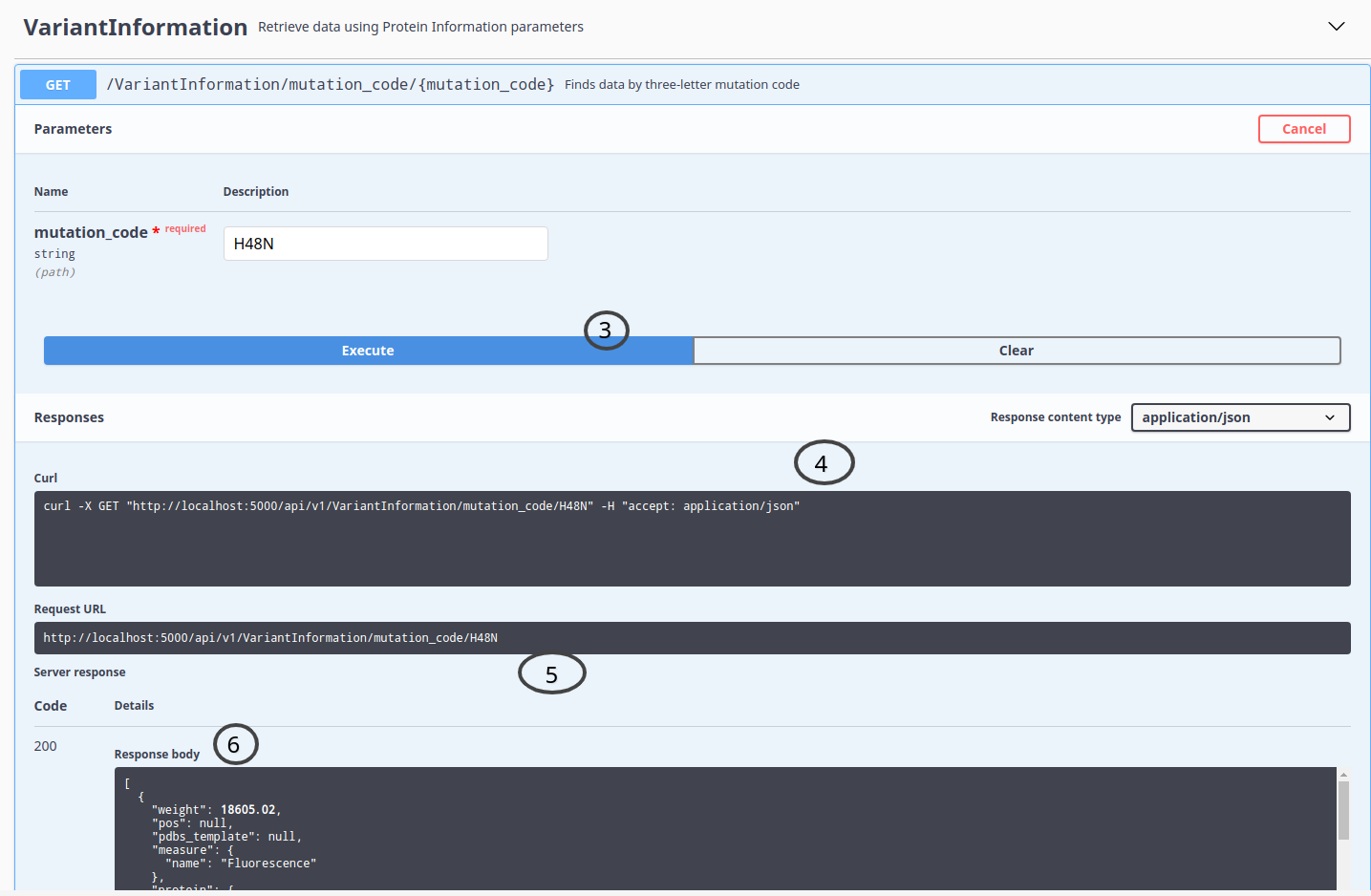
ThermoMutDB API also allows requests by URL.
To perform your URL follow the pattern: API url + name of endpoint + attribute + parameter
To know what variables could be used, check the API documentation form, select an endpoint, copy the GET operation path, and replace variables in brackets to your parameters.
For example, to request data which number of mutations are between 2 and 5 use the URL: http://biosig.unimelb.edu.au/thermomutdb/api/v1/VariantInformation/number_mutations/from/2/to/5
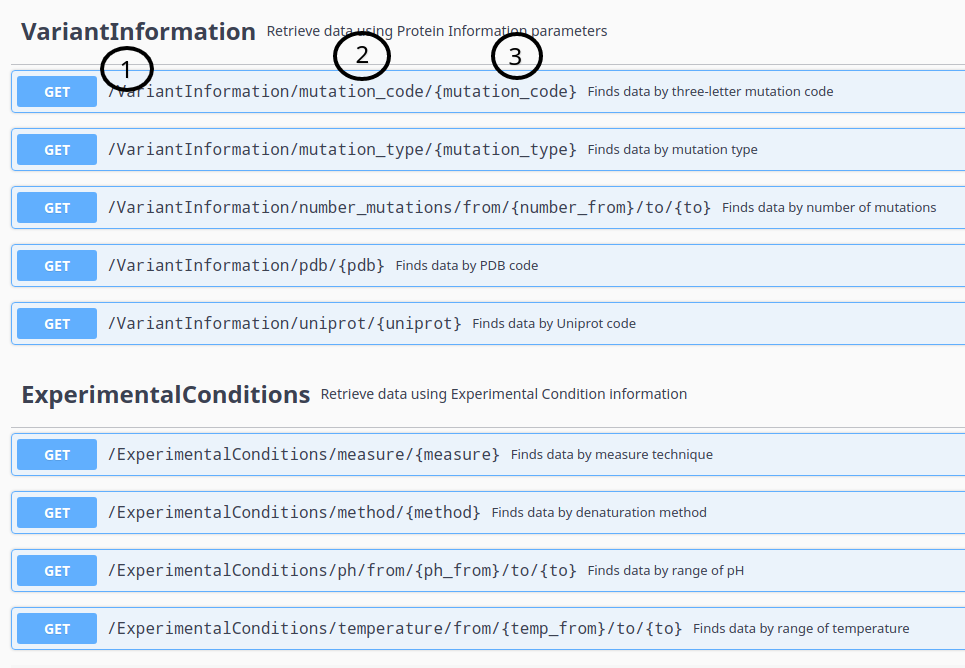
Request by Curl are accepted and needs URL presented in "Request by URL", for example, to request data which have ΔΔG between 1 and 5 use:
curl -X GET "http://biosig.unimelb.edu.au/thermomutdb/api/v1/ThermodynamicData/ddg/from/1/to/5" -H "accept: application/json"
Note: Soon we will provide more flexible criteria to be used in curl and URL requests. All improvements will be published here.
Information content of ThermoMutDB
| Content | Description | Measure* | API attribute |
|---|---|---|---|
|
Protein Information |
|||
|
Protein name |
Protein Name |
- |
|
|
Source |
Protein Organism |
- |
|
|
Uniprot |
Uniprot Code |
- |
|
|
PDB wild |
Protein Data Bank code |
- |
|
|
PDB mutant |
PDB code for mutant (when is available) |
- |
|
|
PDBs template |
PDBs used as a template to model wild_type (when is available and necessary) |
- |
|
|
Length |
Length of sequence |
- |
|
|
Weight |
Molecular weight |
- |
|
|
PIR ID |
Protein Information Resource |
- |
|
|
SWISSPROT ID |
Code of Swiss-Prot (Revised Entries) |
- |
|
|
Mutation |
Three-digits mutation code |
- |
|
|
Mutated chain |
Mutated chain |
- |
|
|
Mutation count |
Number of mutations |
- |
|
|
Experimental Conditions |
|||
|
Temperature |
Experimental temperature |
Kelvin (K) |
- |
|
pH |
Experimental pH |
- |
|
|
Measure |
Experimental techniques for studying protein folding. |
- |
|
|
Method |
Techniques to denature a protein |
- |
|
|
Thermodynamic data |
|||
|
ΔΔG |
Variation of Free Gibbs Energy on the experiment |
kcal/mol-1 |
- |
|
ΔTm |
Variation of Melting Temperature on the experiment |
Kelvin (K) |
- |
|
Structural environment |
|||
|
SST |
Secondary Structure classification |
- |
|
|
RSA |
Relative accessible surface area |
- |
|
|
PHI |
Phi angle value |
- |
|
|
PSI |
Psi angle value |
- |
|
|
Residue Depth |
The average distance of atoms of wild-type residue from the solvent accessible area |
- |
|
|
CA Depth |
The average distance of atoms of CA from the solvent accessible area |
- |
|
|
Relative B Factor |
Temperature factor |
- |
|
|
Substitution matrices scores |
|||
|
Blosum 62 |
BLOSUM 62 matrix score |
- |
|
|
Pam 250 |
PAM 250 matrix score |
- |
|
|
Pharmacophore changes |
|||
|
POS |
Positive |
- |
|
|
NEG |
Negative |
- |
|
|
ACC |
Hydrogen bond acceptors |
- |
|
|
DON |
Hydrogen bond donors |
- |
|
|
ARO |
Aromatic rings |
- |
|
|
SUL |
Sulfuric acid |
- |
|
|
NEU |
Neutral |
- |
|
|
Literature information |
|||
|
Reference |
Publication reference |
- |
|
|
PMID |
Pubmed code of publication |
- |
|
|
DOI |
Digital Object Identifier of publication |
- |
|
|
YEAR |
Year of publication |
- |
|
* If there is.