Help - How to use PLATINUM
Main page
About PLATINUM
PLATINUM is a manually curated, literature-derived database that associates experimental information on changes in protein-ligand affinity with the three-dimensional structures of the complex. The current version of PLATINUM contains structural and affinity data reflecting the effects of mutations on protein-ligand complexes comprising:
- Over 1000 data points (approximately 80% of which are single-point mutations) with protein-ligand affinity data extracted from over 180 peer-reviewed manuscripts;
- More than 200 distinct ligands;
- Approximately 300 distinct PDB structures assigned to 140 different Uniprot entries.
Search page
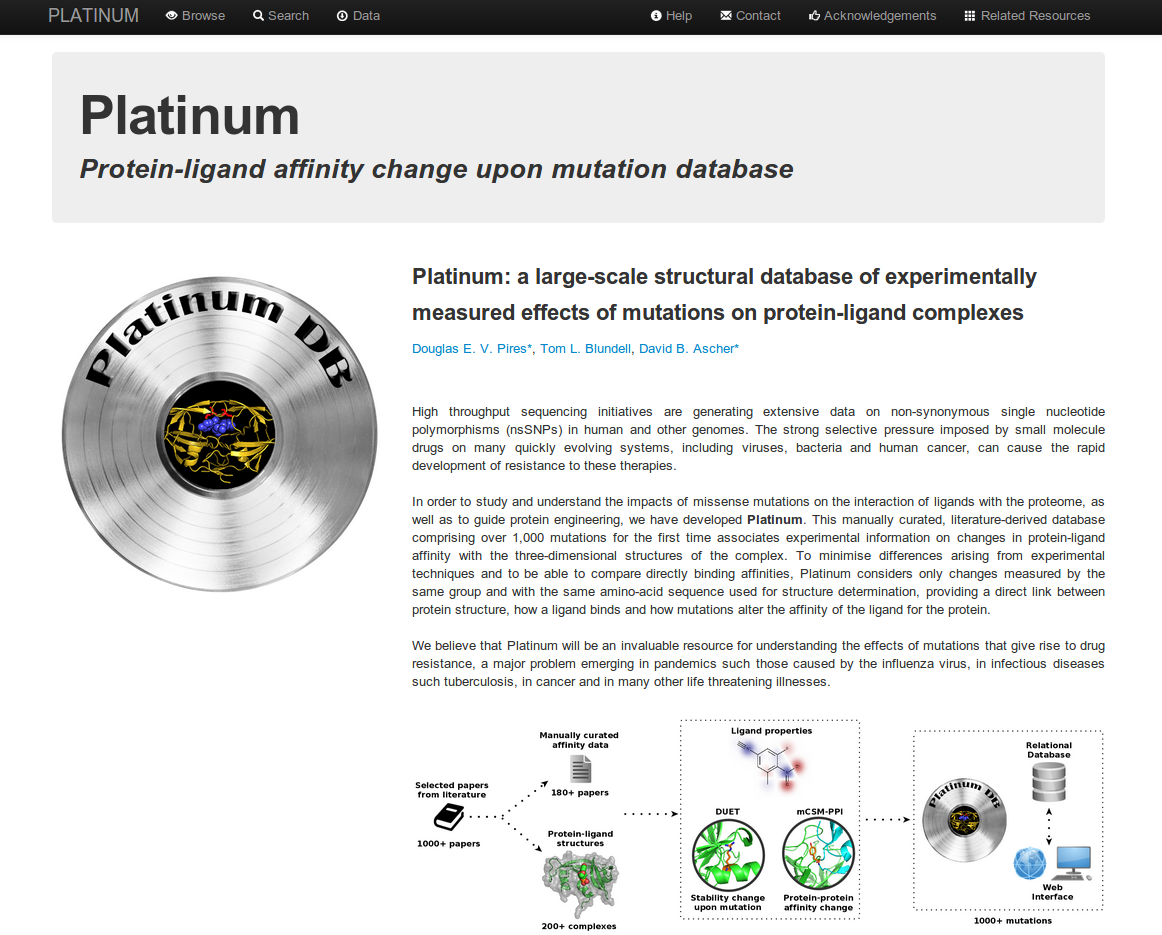
How to query the database:
The complete set of mutations, structural and affinity data in PLATINUM can be browsed by clicking
in (1). Alternatively, users can search (2) for specific categories using the following criterium:
- Ligand type (3), classified according to PDBeChem;
- Organism category (4), which is broadly grouped by phylogenetic kingdoms;
- Mutation effect (5), to select mutations that either decrease or increase protein-ligand affinity;
- Protein classification (6), as assigned by the PDB;
- Experimental method (7), to choose how the affinities were determined;
- Structure resolution range (8), which allows you to select data based upon the resolutions of the complex structures.
- Mutation type (9), which allows you to view only single-point mutations;
- Affinity constant (10), which allows you to view only KD measurements;
Database page
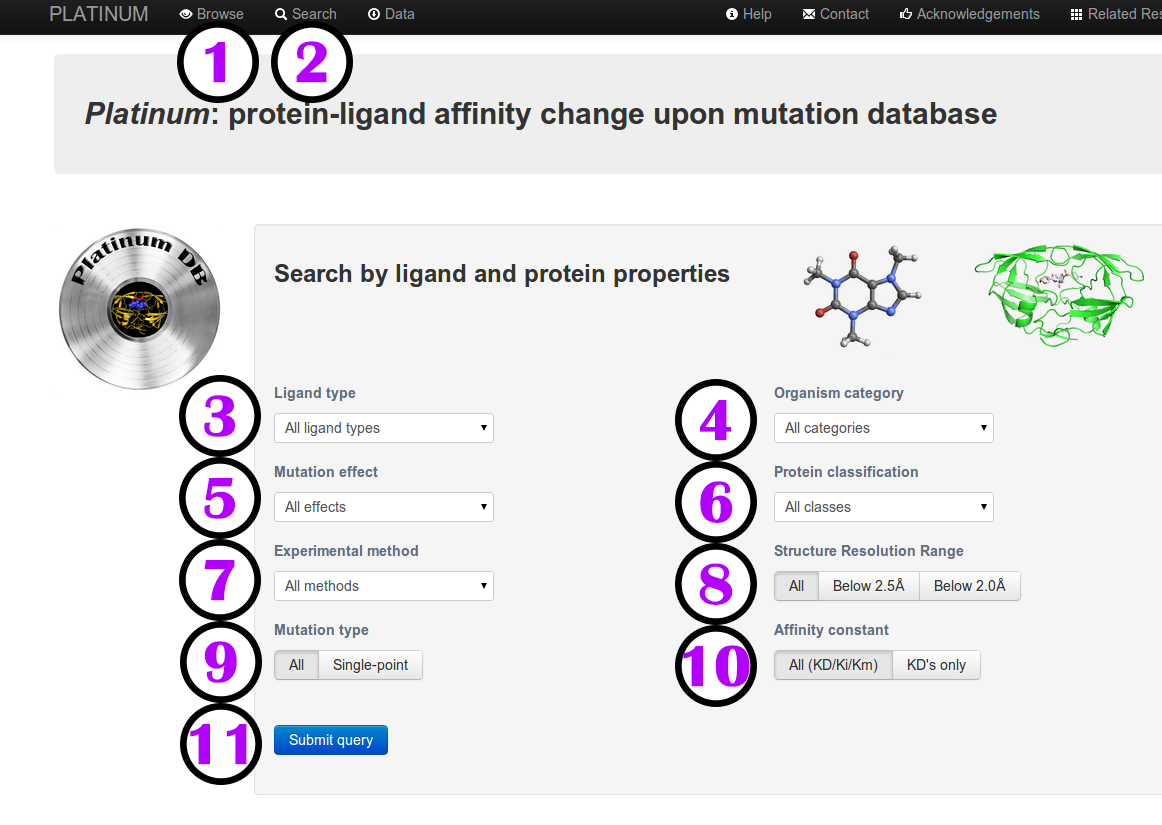
Accessing the database
After clicking on Browse or performing a Search the database entries will be displayed in a tabulated
format on this page.
The rows are coloured according to the effect of the mutation upon the binding affinity of the ligand.
Red indicates that the mutant has a weaker affinity than the reference, while blue indicates that the
mutant has stronger affinity.
Columns descriptions are available by hovering your cursor over the column headings (tooltip function).
The headings of the hidden columns are coloured according to property they are linked to.
The information that is displayed can be adjusted according to your interests.
Additional information regarding the following aspects can be shown or hidden:
- Residue properties (1), when reference structure is available;
- Ligand properties (2);
- Affinity experimental details (3) and
- Protein structural information (4).
The number of records displayed per page can be selected at (5) and the filtered results can be downloaded as a comma-separated .csv file (6), which can be easily imported by any spreadsheet program (e.g., LibreOffice, Microsoft Excel, Numbers, etc.) or any text editor.
Database page
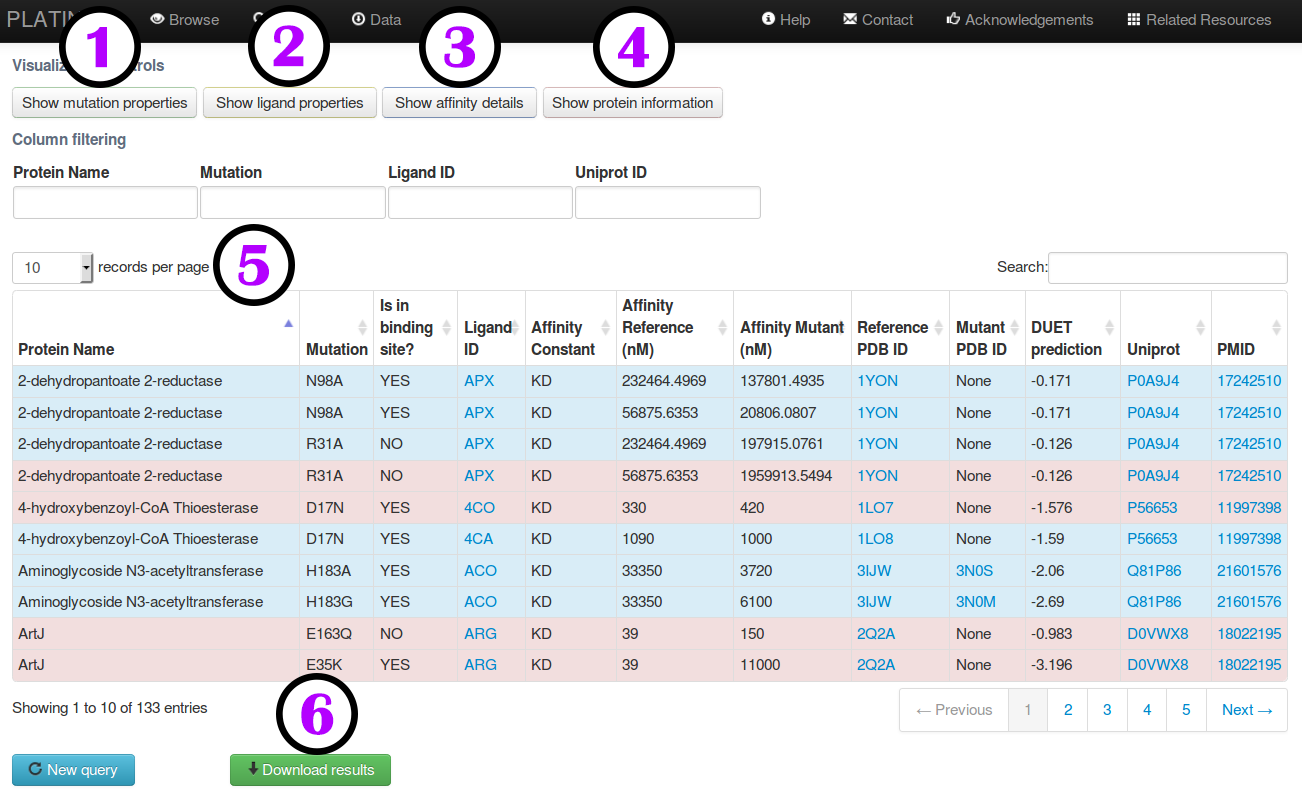
Filtering query results
Once a query result is displayed users can select subsets of mutations of interest by either using a
global search (1) or a field-specific filtering tools.
The supported fields are:
- Protein name (2);
- Mutation (3);
- Ligand ID (4) and
- Uniprot ID (5).
- Mutations affecting GDP binding (Ligand ID: GDP);
- For the C. elegans GDP-fucose protein O-fucosyltransferase 1 (Uniprot ID: Q18014) and
- Using a regular expression (Mutation: A$, which denotes "last character of the field is A") to filter only alanine scanning mutations.
Data page
Downloading the database
The data page (1) allows users to download the complete set of information available in PLATINUM:
- You can download PLATINUM as a single flat-file (comma-separated) by clicking on (2).
- Additionally, you can also download the full set of processed protein-ligand complexes in PDB format (3).
Contact page
Getting in touch
In case you experience any trouble using PLATINUM or have any suggestions or comments, please do not hesitate in contacting us (1) either via e-mail (2) or through the online form (3).