About
What is the Missense Tolerance Ratio?
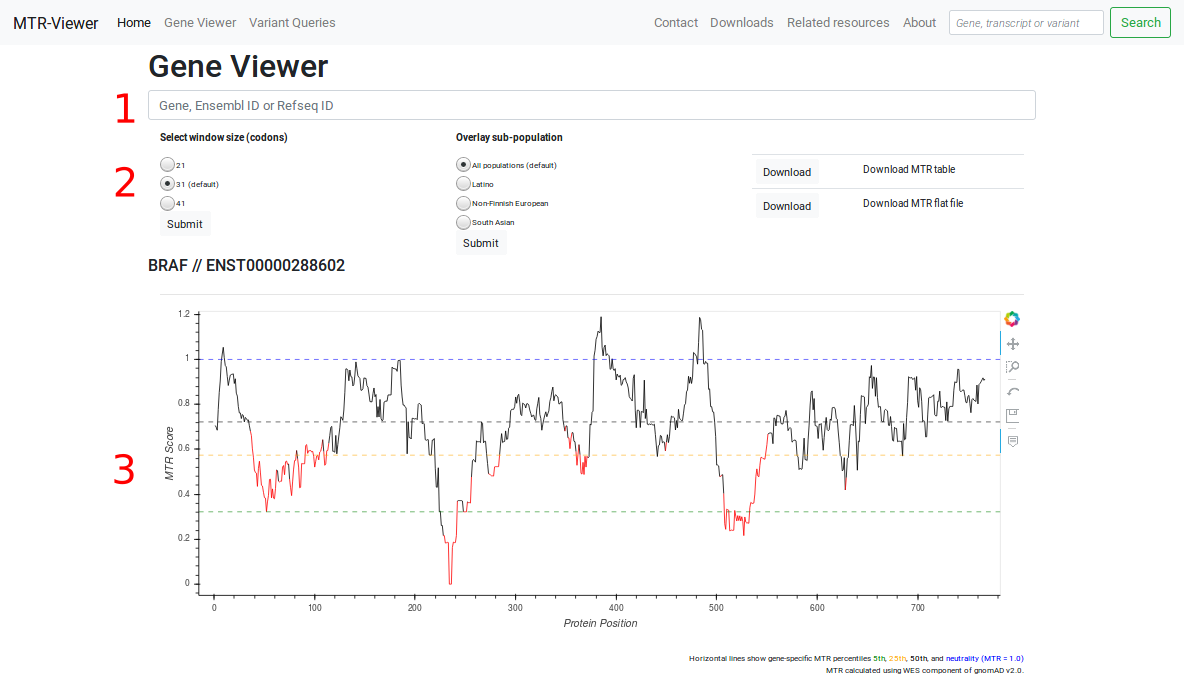
The MTR aims to quantify the amount of purifying selection acting specifically on missense variants in a given window of protein-coding sequence. It is estimated across sliding windows of 31 codons (default) and uses observed standing variation data from the WES component of gnomAD / the Exome Aggregation Consortium Database (ExAC), version 2.0 (http://gnomad.broadinstitute.org).
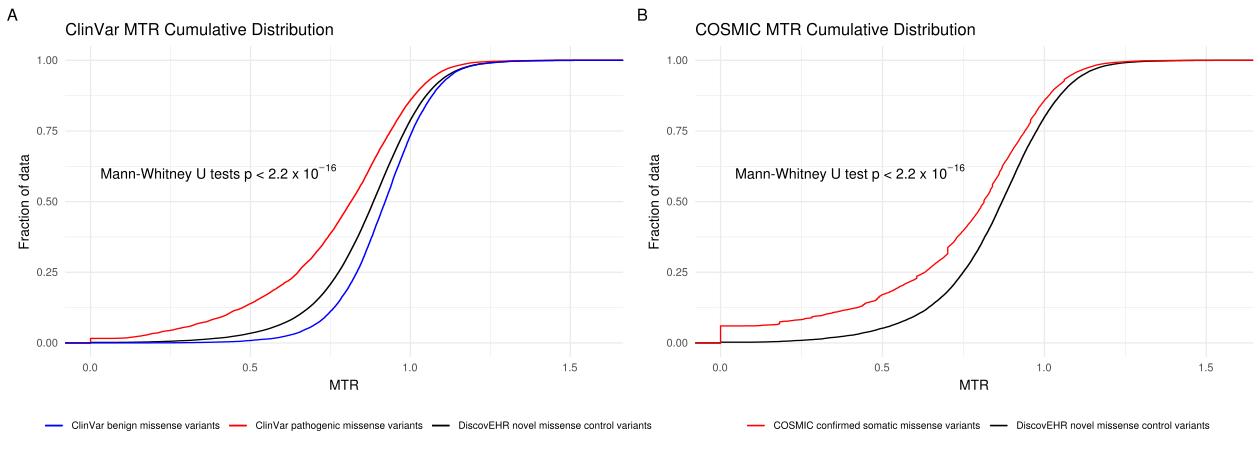
Distribution of MTR scores for known disease variants compared to background. (A) Cumulative distribution of MTR scores for ClinVar pathogenic variants (red), ClinVar benign variants (blue) and DiscovEHR novel missense control variants (black). (B) Cumulative distribution of MTR scores for COSMIC somatic missense variants (red) compared with DiscovEHR novel missense control variants (black).
How are the MTR scores calculated?
Please refer to the associated publication, Traynelis J.,* Silk M.,* Wang Q., Berkovic S.F., Liu L., Ascher D.B., Balding D.J., Petrovski S. (2017). Optimizing genomic medicine in epilepsy through a gene-customized approach to missense variant interpretation. Genome Research (2017).
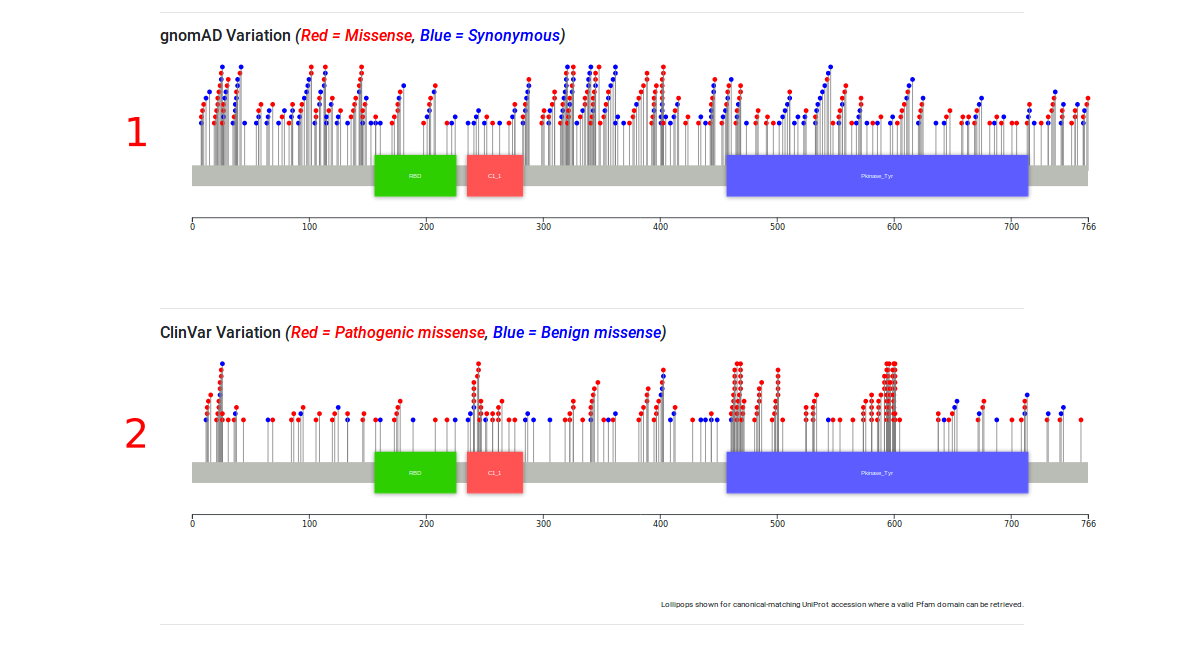
Lollipop Plots of Variation
Lollipop plots are given for the gnomAD variation for a given gene, where there i sa valid HGNC symbol and Pfam domain representation for that symbol. If the HGNC symbol matches to a gene in ClinVar's set of disease-associated genes, a lollipop plot is given for the non-ambiguous benign and missense variants found.
These are adapted from the GitHub project here: https://github.com/pbnjay/lollipops
Can I access the MTR scores via any batch processes?
The MTR scores are available via the Variant Effect Predictor (VEP) using a plugin at https://www.ensembl.org/info/docs/tools/vep/script/vep_plugins.html. This can be used to annotate variants with the MTR scores.
They can also be accessed using an API to query variants using a web-browser http://biosig.unimelb.edu.au/mtr-viewer/api/search or via command line using curl -i http://biosig.unimelb.edu.au/mtr-viewer/api/search, where "search" allows for identical query formatting as in the queries page.
Querying via the API returns a JSON object with the same columns each time, and can be used for easy pipeline integration.
How do I use the MTR-Viewer?
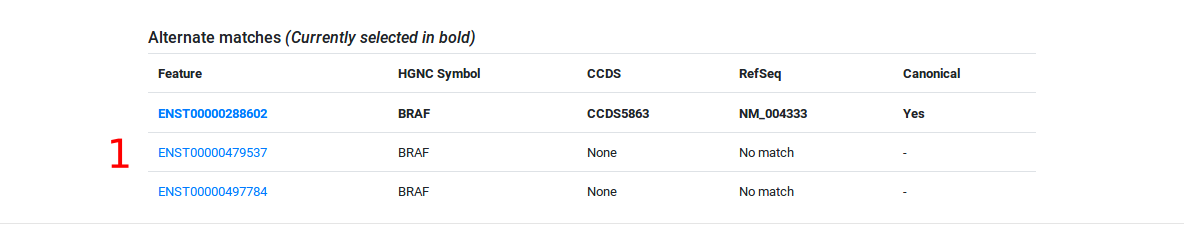
Enter a gene symbol (HGNC), Ensembl transcript ID (v75) or corresponding Refseq transcript ID into the search bar. The viewer will display the gene name and Ensembl Transcript ID. If a gene symbol (HGNC) is supplied, the viewer will default to a canonical transcript of our choosing. Alternate transcripts are listed below the viewer.