1.Submission Page
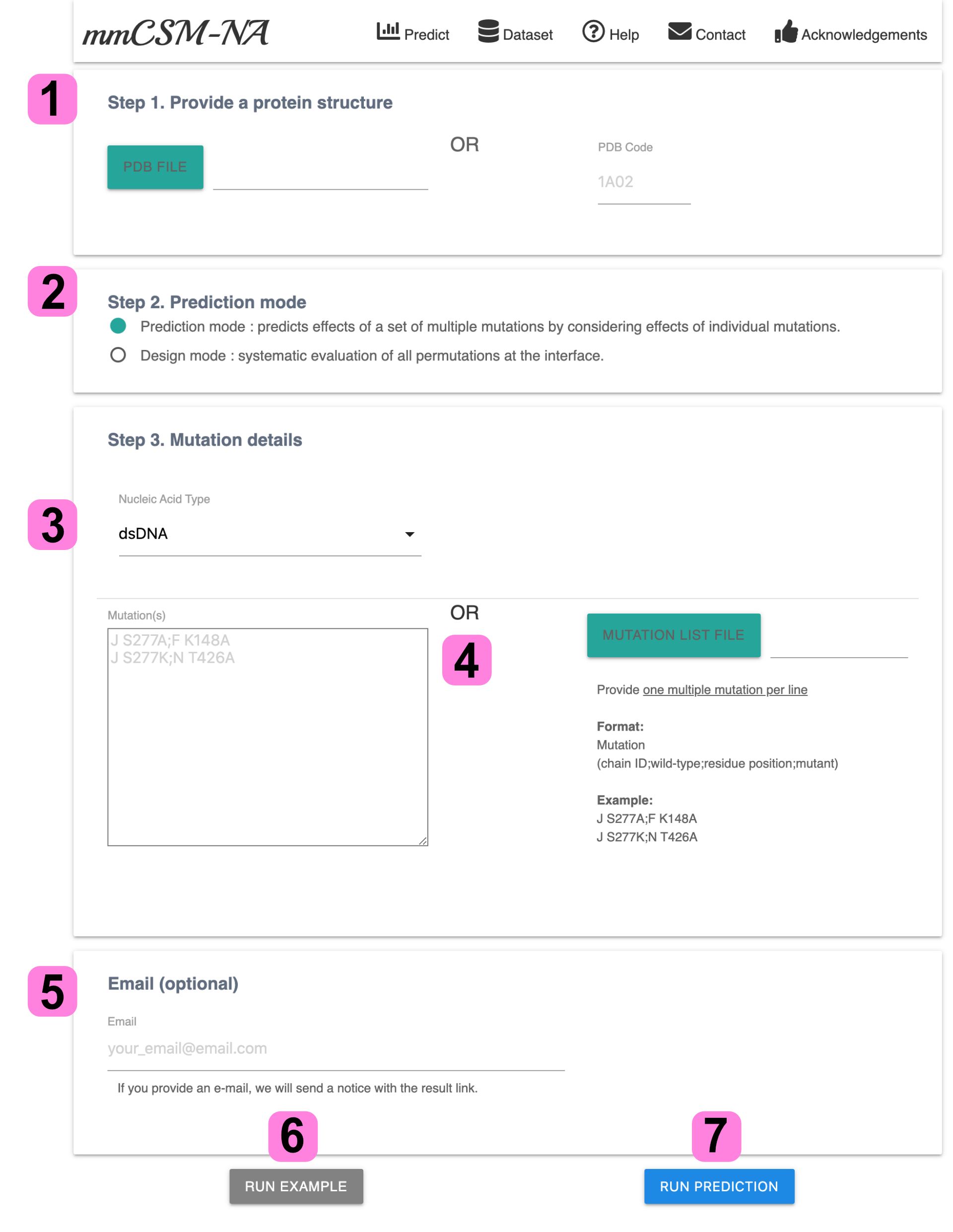
1. Provide a nucleic acid - protein complex by either of PDB file or PDB code.
2. Choose prediction modes between Prediction and Design based on the purpose of the prediction.
3. For Prdiction Mode, choose one of nucleic acid types and
4. the mutation details can be given by typing in input textarea or uploading a mutation list file (support more than one multiple mutation).
The Design Modes automatically allocate the binding interface residues between nucleic acids and proteins for design mode predictions.
5. By providing an e-mail address, user can a notification email, after job completion.
6. To visit example pages for each mode, users have to select a prediction mode from step 2 and hit 'Run Example' button.
7. Click RUN PREDICTION to get your prediction result.
2.Result Page- Prediction Mode
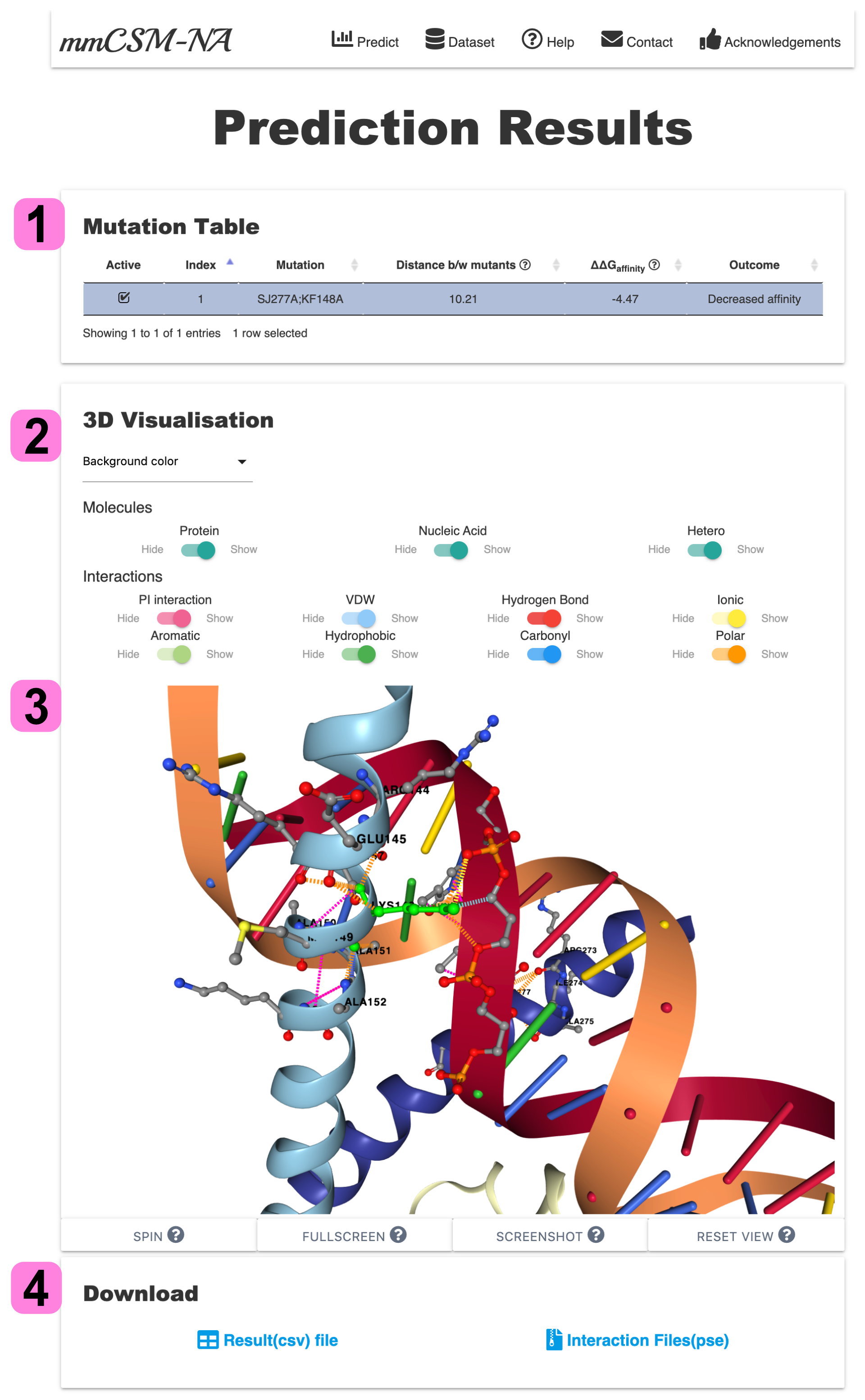
The result page of Prediction Mode is divided into three sections:
1. The Mutation Table section gives predicted ∆∆G and distance among mutants. If the mutation is more than two per case, the distance bw mutants shows distances between the centroid of each mutant and each Calpha carbon.
Users can browse mutations in the wild-type structure via 3D Visualisation ticking a checkbox of each multiple mutation from Mutation Table.
2. The 3D Visualisation provides wild-type atomic interactions of a given multiple point mutation. Using control panel, users are able to change the representation of structure.
3. Each molecule and interaction (the colours are matched with the dashed-line in 3D viewer) can be turned on/off using switches.
4. The mutation and interaction information can be downloaded as Pymol-session(PSE) and CSV file, respectively.
3.Result Page- Design Mode
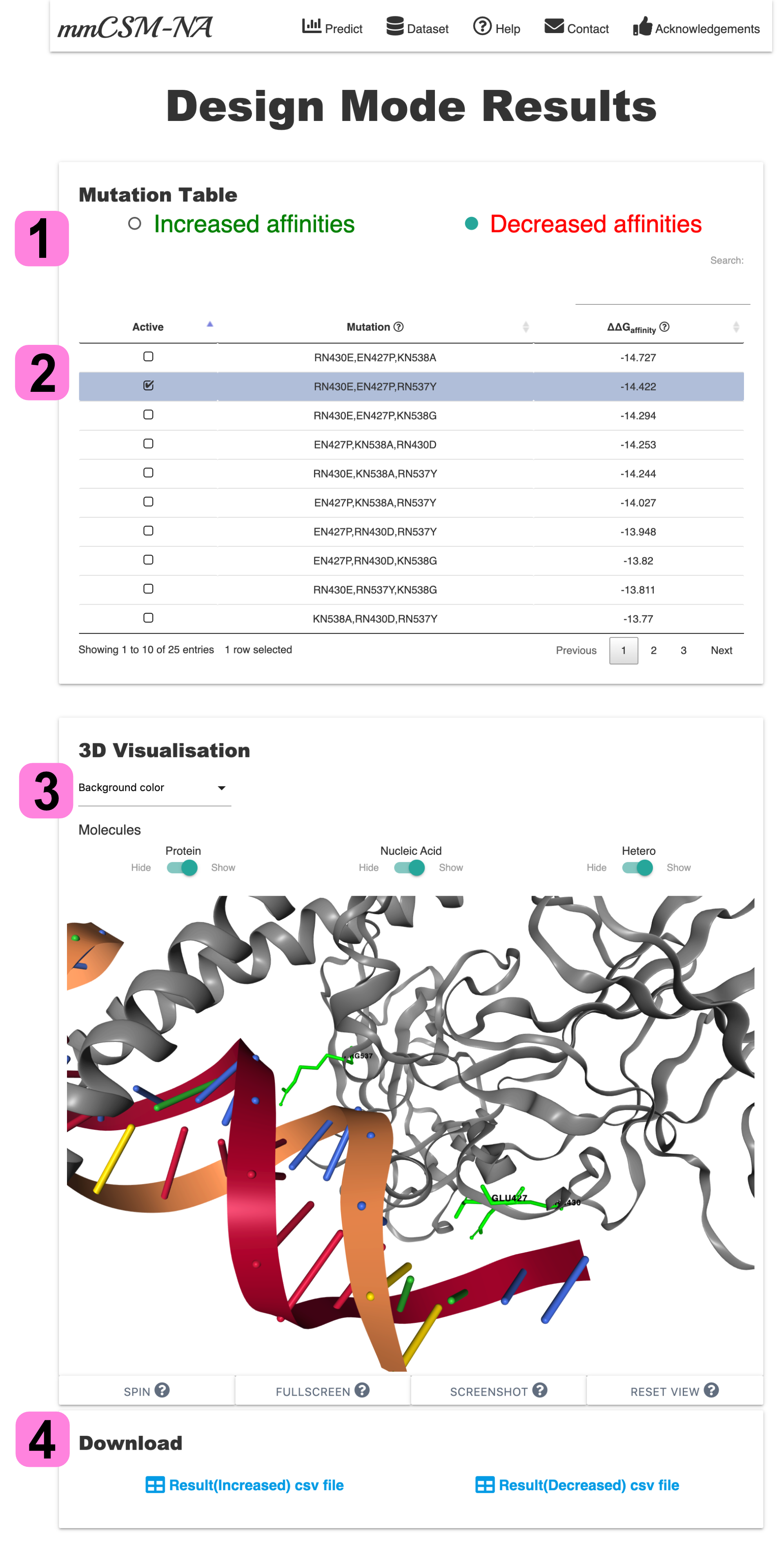
1. Mutation Table shows the top 100 destabilising/stabilising mutations for nucleic acid and protein interface residues. The predicted ∆∆G and mutation information are shown in the table. Users can select stabilising or destabilising datatable.
2. Once a datatable is selected, users can browse or search for information in the datatable.
3. If user selects a mutation(row) in the table, 3D Visualisation highlights the location of mutations in green. The representation of biomolecules can be turned on/off through the control panel.
4. Users can download both stabilising/destabilising datatable in CSV file separately.