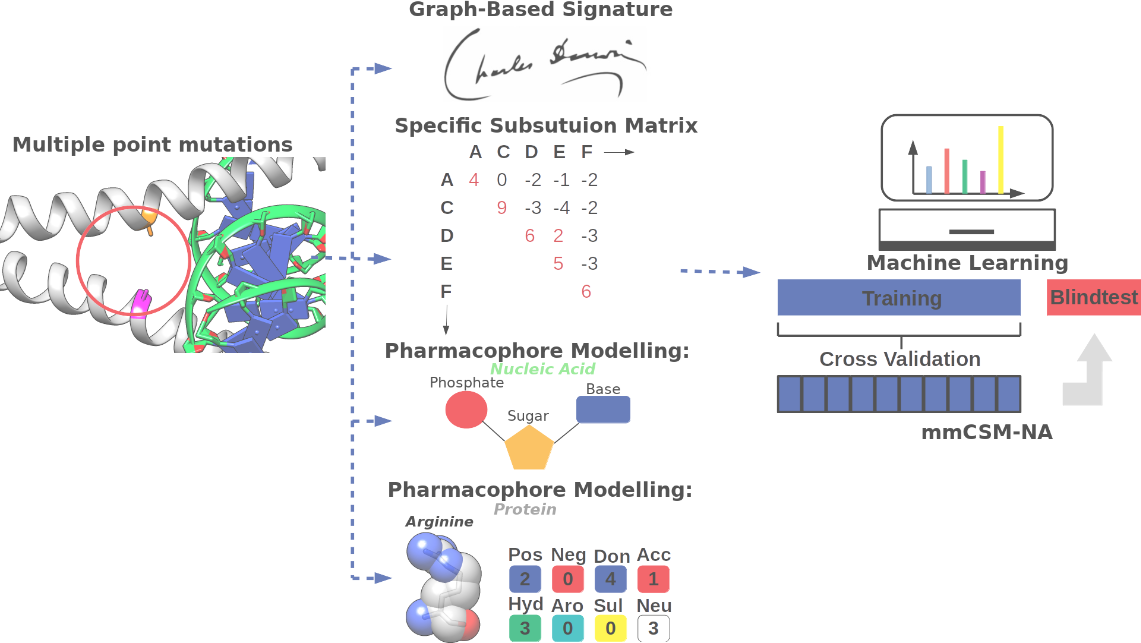
mmCSM-NA: accurately predicting effects of single and multiple mutations on protein-nucleic acid binding affinity
The interaction between proteins and nucleic acids plays pivotal roles
throughout transcription,translation, and gene regulation. We previously pioneered the development of computational
approaches to look at the effects of single point missense mutations on protein-nucleic acid interactions (mCSM-NA and
mCSM-NA2).
While these and subsequent methods have been widely used, there has been a pressing need for the
ability to assess the effects of more complex mutations, as most protein and system engineering efforts require
the introduction of >2 independent missense mutations.
Here, we have adapted our well validated concept of graph-based signatures to better capture the
changes at the protein and nucleic acid interface in order to more accurately predict the effects of both single and multiple
point mutations on protein-NA binding affinity: mmCSM-NA. To the best of our knowledge, this is the first method of
its kind,and represents a significant advance upon previously developed tools.