mCSM-membrane: How to use
Tip! mCSM-membrane can be used with homology models. Try automated comparative modelling online with ModWeb and SwissModel.
Tip! Try using the key arrows in your keyboard or the buttons below to switch among the tips below.
Main page
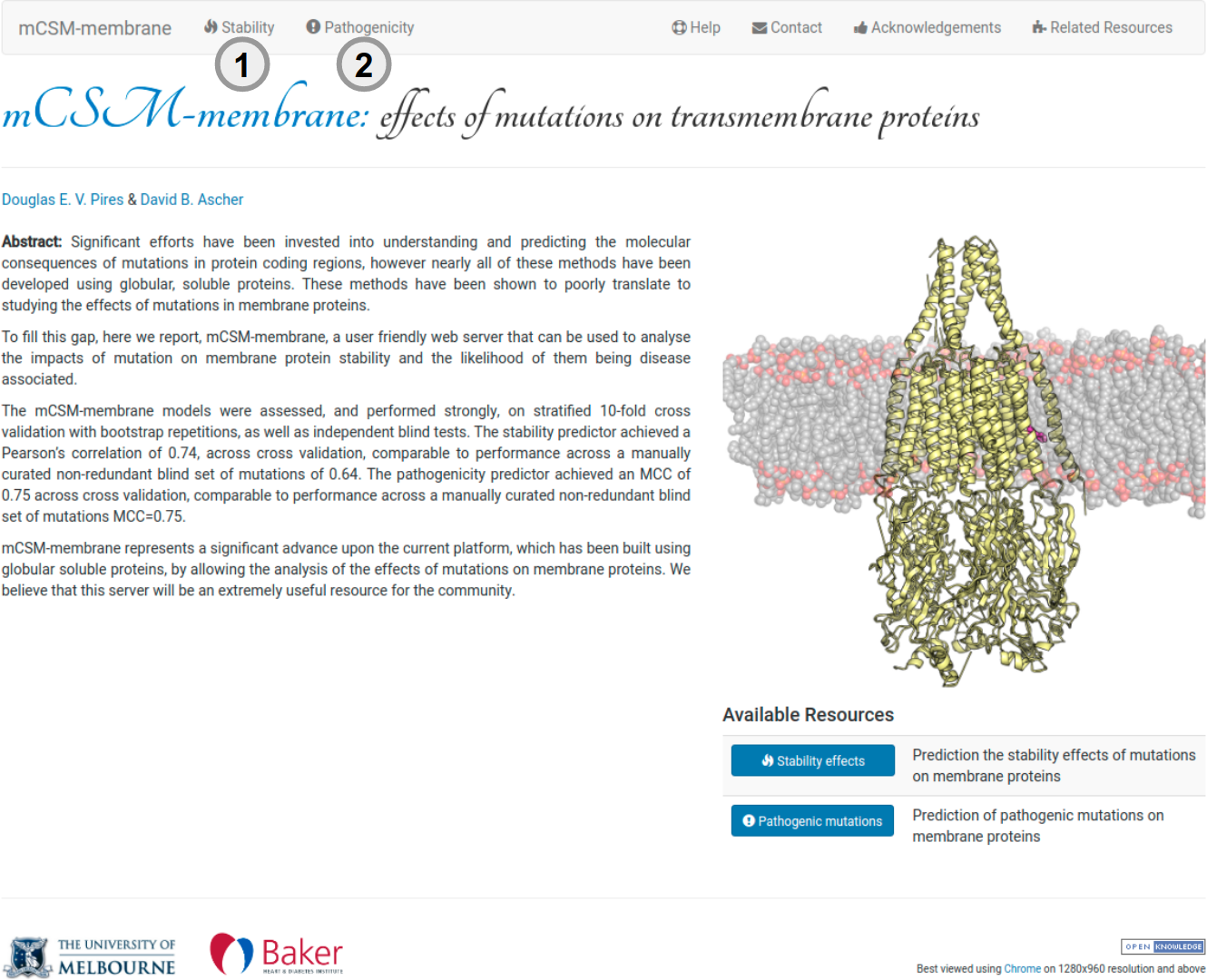
About mCSM-membrane
mCSM-membrane is a user friendly web server that can be used to analyse the impacts of mutation on the stability of membrane protein and the likelihood of them being disease associated. It is a machine learning approach that uses graph-based structural signatures to train and test predictive models. mCSM-membrane represents a significant advance upon our current predictive platform, which has been built using globular soluble proteins, by allowing the analysis of the effects of mutations on membrane proteins. To analyse the effects of mutation on stability click in (1) in the navigation or in (2) to predict whether mutations are pathogenic or benign.
Submission page
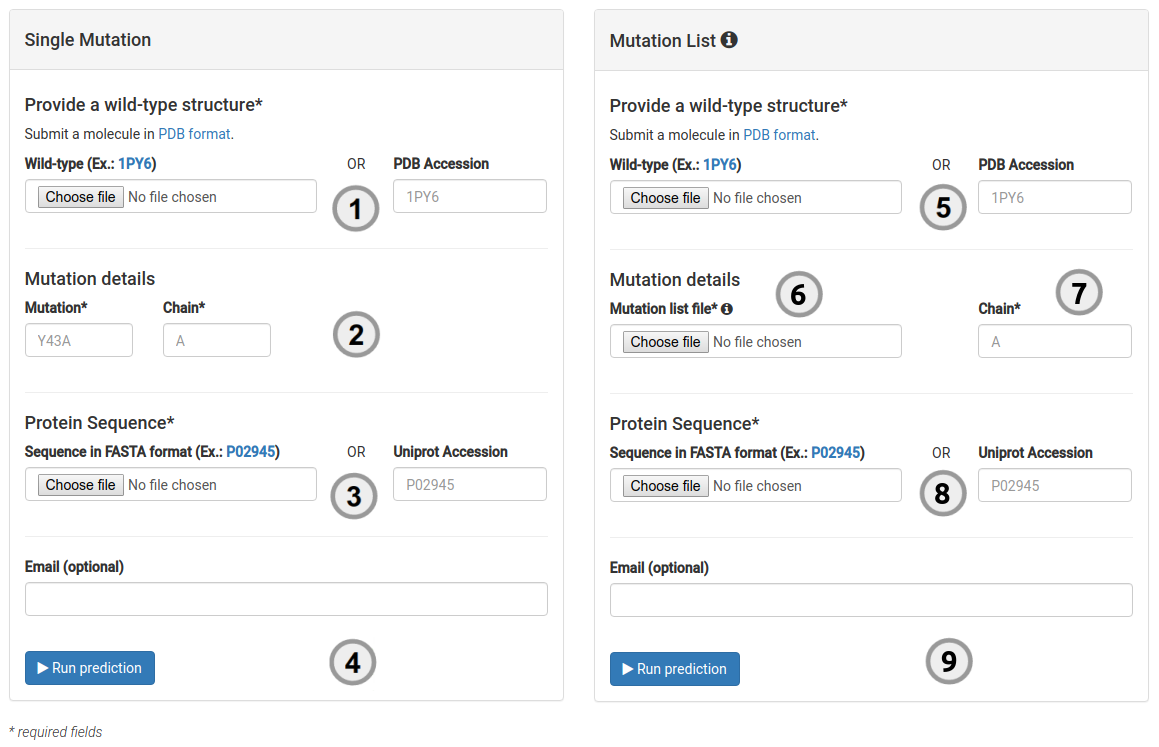
How to run a prediction
To run a single prediction for either stability effects or the likelihood of a mutation being pathogenic:
- Provide the structure of the wild-type protein, which must comply with the PDB format by either providing a PDB accession code or by uploading your own structure (1).
- A single mutation to be analysed should be provided (2) via a mutation code and chain ID. A mutation code consists of wild-type code, residue position and mutant code (using the one letter amino acid code). Residue position must be consistent with the PDB file. The chain for the mutation code must also be provided.
- Provide the UNIPROT accession code for your protein protein (3).
- You are then ready to submit your query for analysis (4).
- Provide the structure of the wild-type protein, which must comply with the PDB format by either providing a PDB accession code or by uploading your own structure (5).
- A file with a list of mutations to be analysed should be provided (6), with one mutation per line, following the aforementioned mutation format. The chain must also be provided and consistent with the PDB file (7).
- Provide the UNIPROT accession code for your protein protein (8).
- You are then ready to submit your query for analysis (9).
Results page
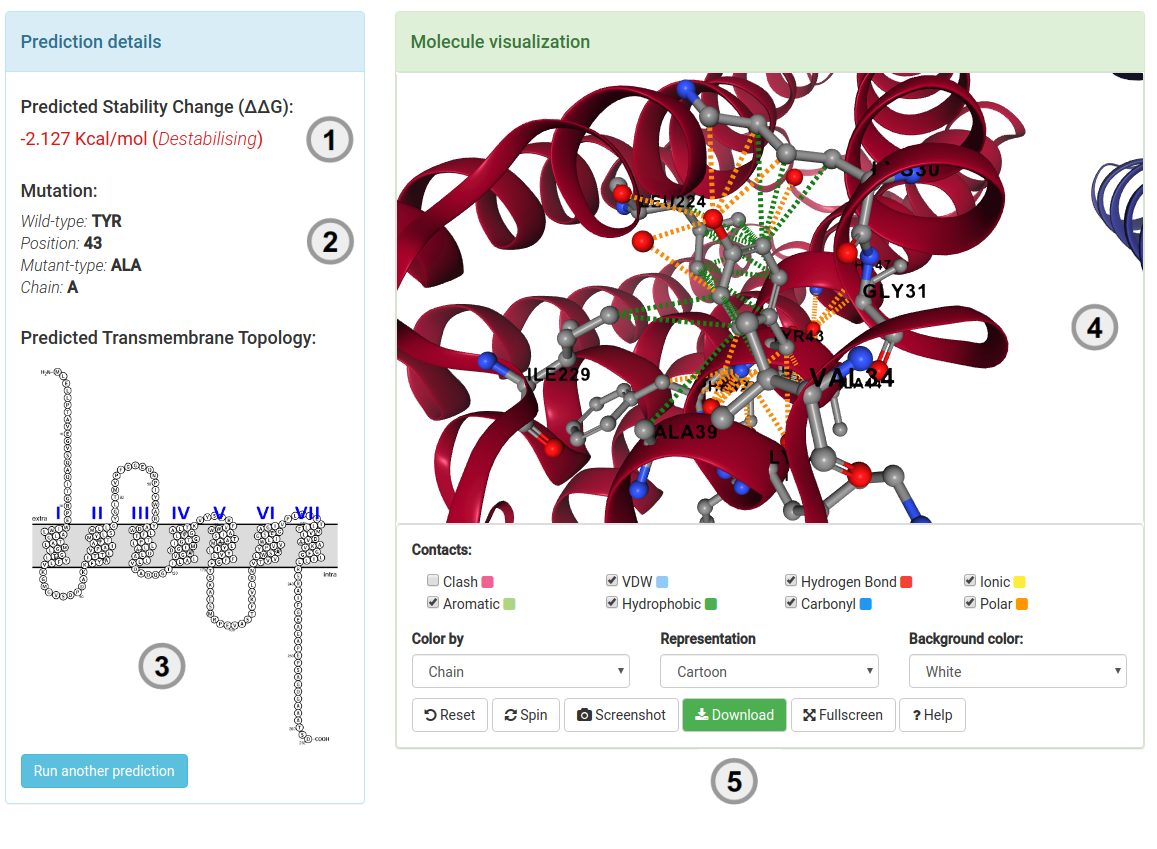
Results - Single mutation
Your results (1) for a single mutation will be displayed once computations are completed. For predicting effects on stability,
the results will display the predicted change in stability upon mutation (ΔΔG in Kcal/mol). A negative value (and red writing) corresponds
to a mutation predicted as destabilising; while a positive sign (and blue writing) corresponds to a mutation
predicted as stabilising.
For predicting pathogenicity, mutations are labeled either as Pathogenic (in red writing) or Benign (in blue writing).
Complementary information also displayed include:
- A summary of the mutation is presented (2) highlighting the wild-type residue, position number and chain.
- A depiction of the predicted transmembrane topology for the provided protein (3) is shown.
- The protein residue environment can be visualised directly from the server (4). Several options are provided for customization of the molecule. Mouse controls are also enabled.
- (5) allows users to download a pymol session of the mutated residue and its interactions.
Results page
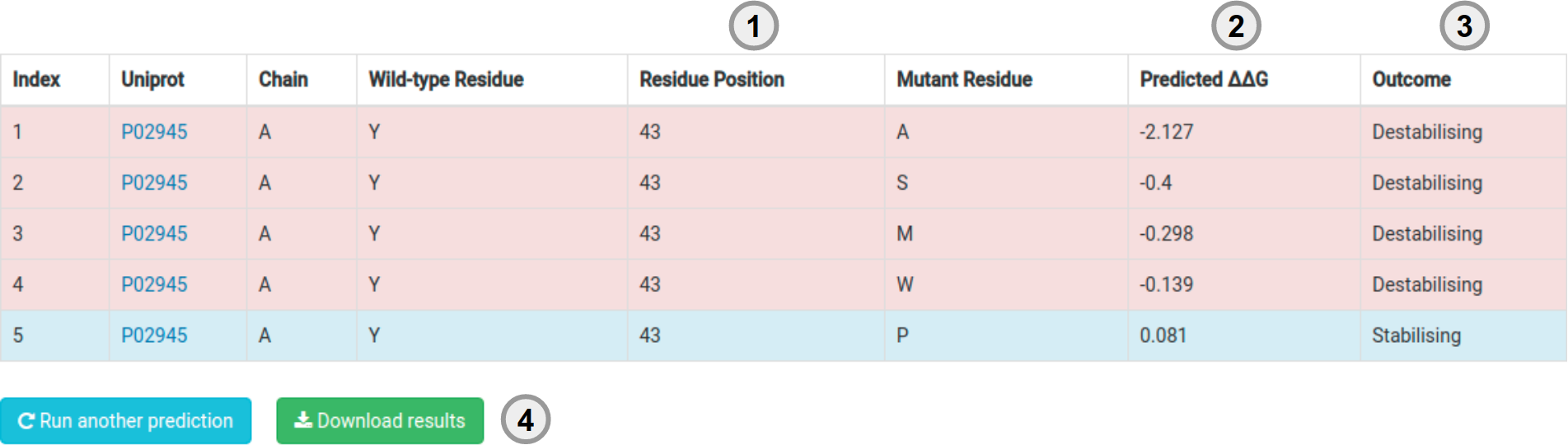
Results - List of Mutations
Your results for a list of mutations will be displayed in a table format with the following information:
- Mutantion information (1).
- The predicted effect (either in stability as a ΔΔG or a pathogenicity prediction) (2) and the predicted outcome (3).
Contact page
Getting in touch
In case you experience any trouble using mCSM-membrane or have any suggestions or comments, please do not hesitate in contacting us either via e-mail (1) or through the online form (2) .