epitope1D: Accurate taxonomy-aware b-cell linear epitope prediction
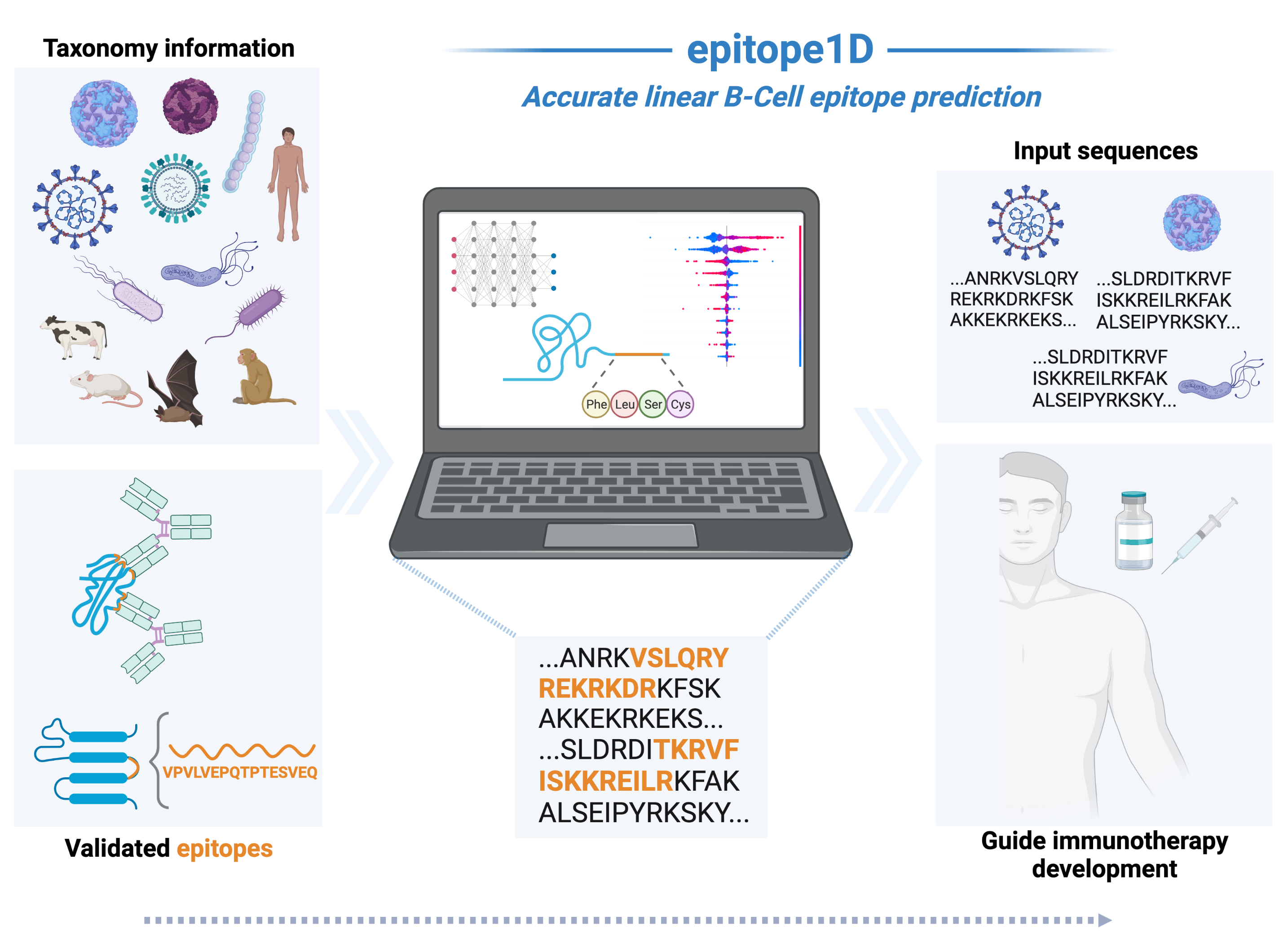
The ability to identify B-cell epitopes is an essential step in vaccine design, immunodiagnostic tests, and antibody production. Several computational approaches have been proposed to identify, from an antigen protein or peptide sequence, which residues are more likely to be part of an epitope, but have limited performance on relatively homogeneous data sets and lack interpretability, limiting biological insights that could otherwise be obtained.
epitope1D, an explainable machine learning method capable of accurately identifying linear B-cell epitopes, leveraging two new descriptors: a graph-based signature representation of protein sequences and the Organism Ontology information. Our model achieved Areas Under the ROC curve of up to 0.935 on cross-validation and blind tests, demonstrating robust performance.
A comprehensive comparison to alternative methods using distinct benchmark data sets was also employed, with our model outperforming state-of-the-art tools.