DockNet is a valuable resource for predicting contact residue probability between two protein partners.
Running predictions
Users are required to provide structures for two protein partners by providing valid PDB accession codes (2) or via uploading files in PDB format (2).
For structures with multiple proteins, users may select specific monomers by providing a chain identifier (3), otherwise the server will automatically select the first protein present on the input structure
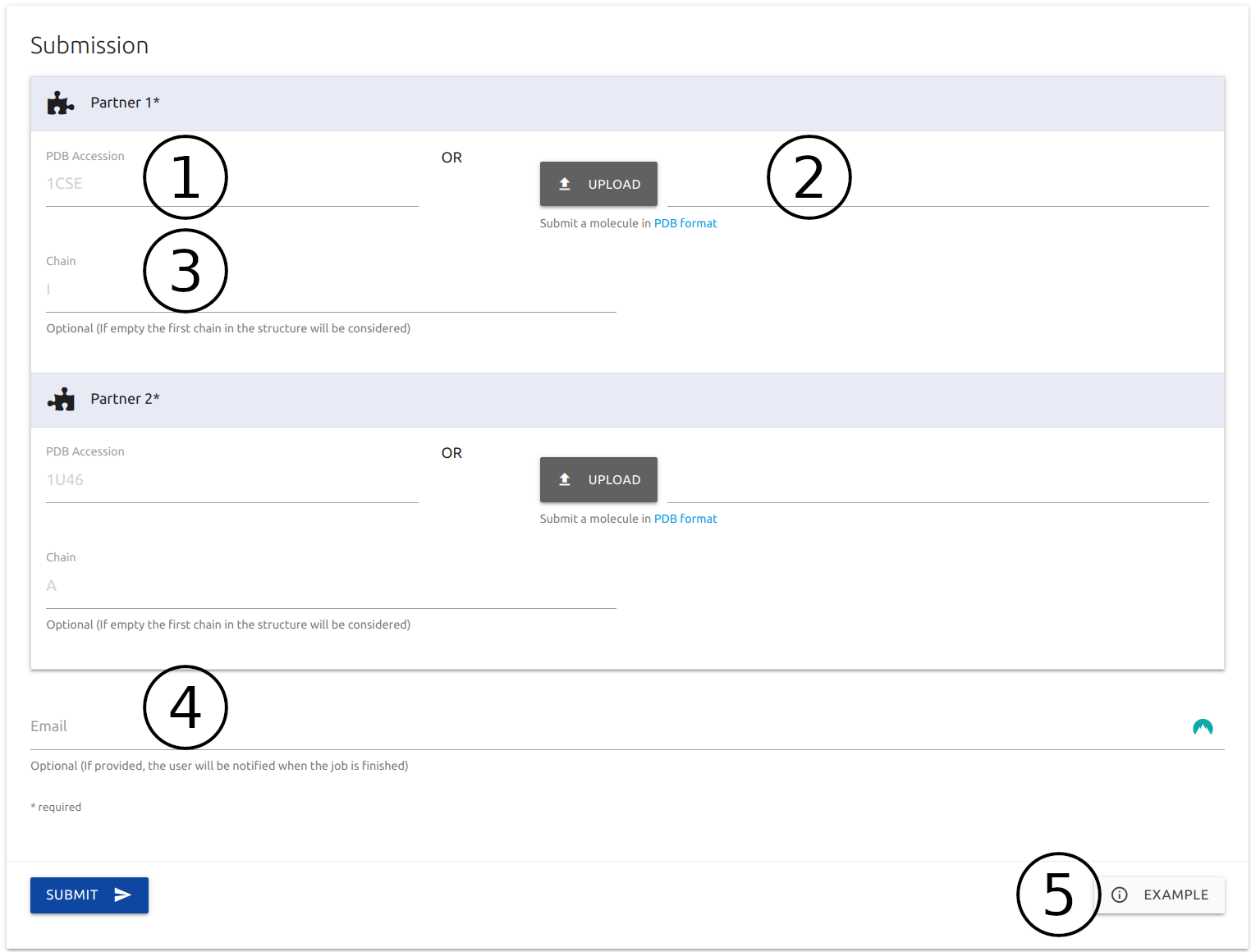
If provided (4), an email will be sent to the user after the submission is processed.
A button for an example results page is available at the bottom of the form (5)
Results
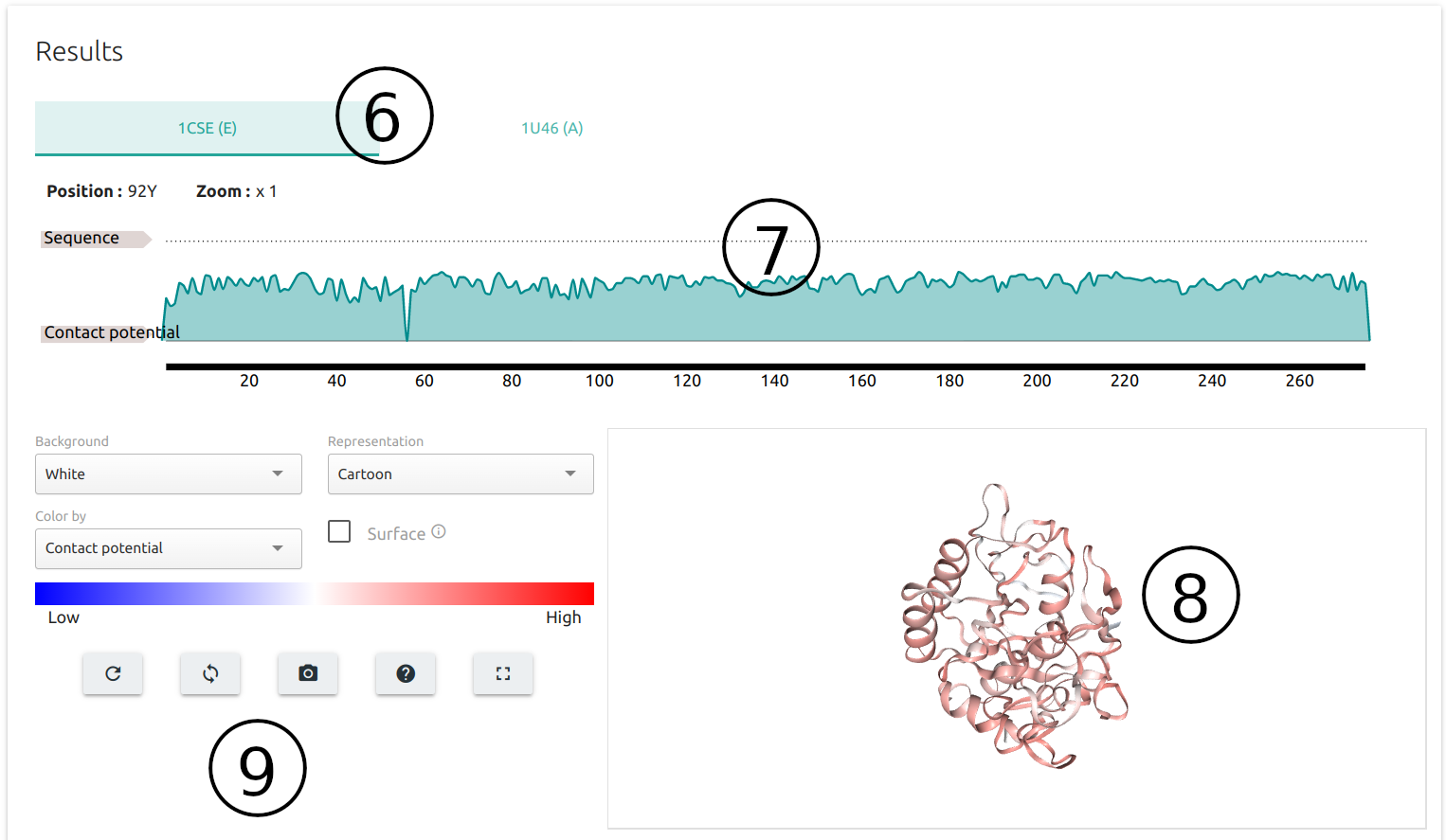
Results are summarised for each protein partner on separate tabs (6), where predictions are:
- averaged per residue and mapped onto the protein sequence using the FeatureViewer component (7)
- and the input 3D structure via an interactive viewer built using NGLviewer (8)
- a set of controllers and action buttons are available for customising the 3D viewer (9).
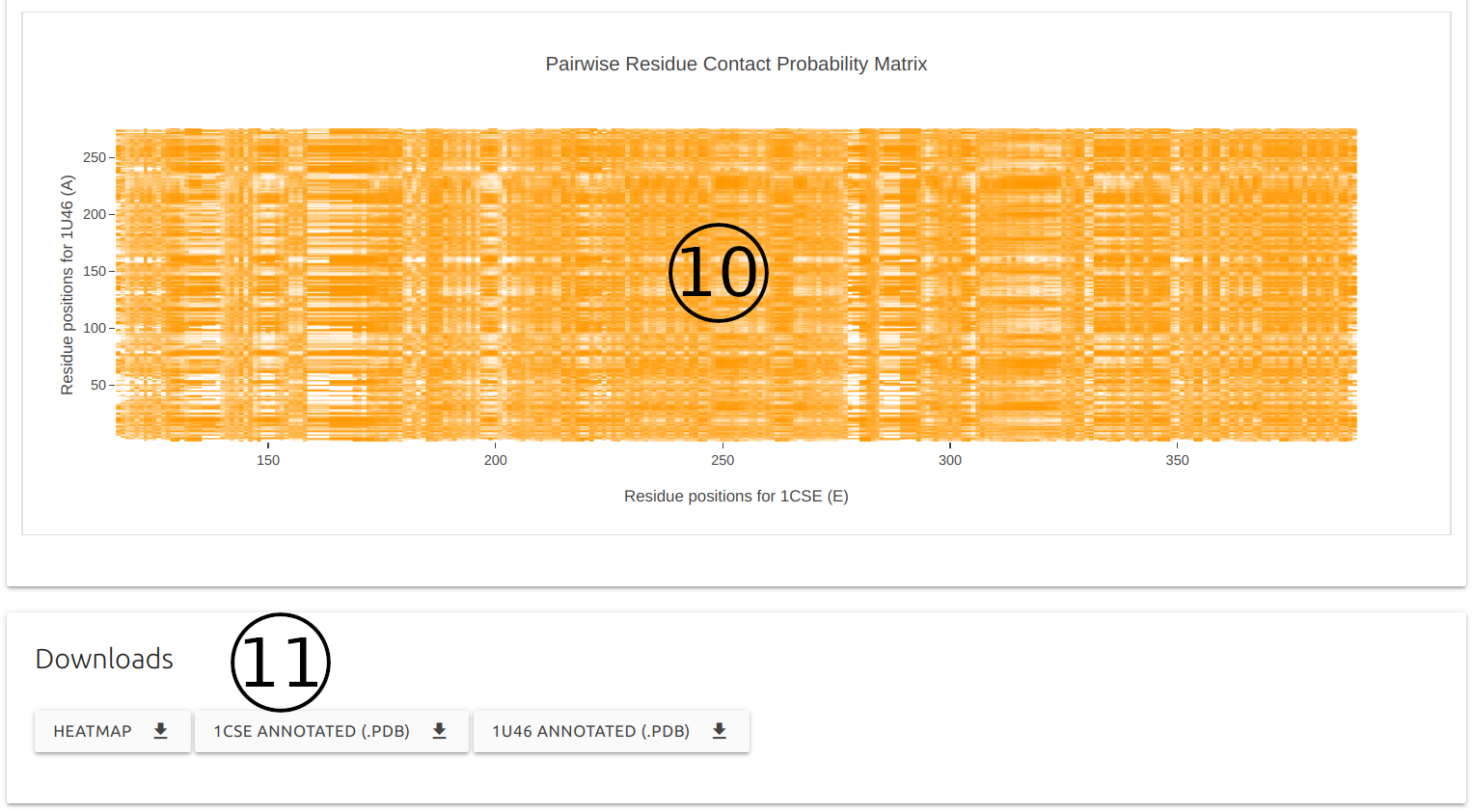
A pairwise residue contact probability matrix is also displayed to help users compare contact potentials between the input proteins (10).
PDB structures for both partners with predicted probabilites for each residue annoted on the b-factor columns are available for download, as well as the pairwise contact matrix as a comma separated file (CSV) (11).
Contact us
In case you experience any troube using DockNet or if you have any suggestions or comments, please do not hesitate in cantacting us either via email or via our Group website.
If your are contacting regarding a job submission, please include details such as input information and the job identifier