DockNet High Throughtput PPI Contact Prediction Using Geometric Deep Learning
Nathan Williams, Carlos H.M. Rodrigues, Jia Q. Truong, David B. Ascher & Jessica K. Holien
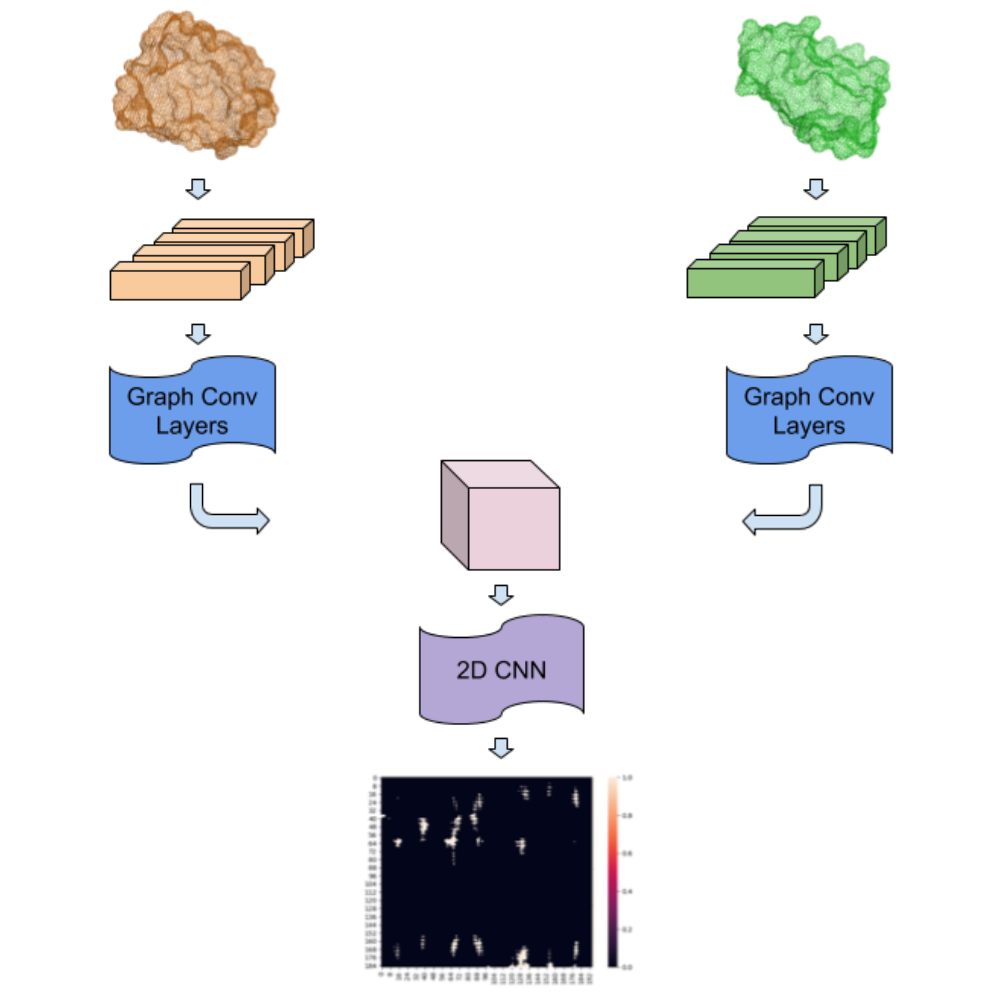
Abstract: Over 300,000 PPI interaction pairs have so far been identified in the human proteome and targeting these PPI interactions is fast becoming the next frontier in drug design. Predicting PPI interaction sites, however, is a challenging task that traditionally requires computationally expensive and time-consuming docking simulations. A major weakness of modern protein docking algorithms is the inability to account for protein flexibility, which ultimately leads to relatively poor results. Additionally, most algorithms do not incorporate evolutionarily conserved sequence features, which can help pinpoint interfaces where this information is unknown. In this manuscript we propose DockNet, an efficient siamese graph-based deep neural network method which predicts contact residues between two interacting proteins. Unlike other methods that only utilise a protein’s surface or treat the protein structure as a rigid body, DockNet incorporates the entire protein structure and places no limits on protein flexibility during an interaction. Predictions are modelled at the residue level, based on a diverse set of input node features including residue type, surface accessibility, residue depth, secondary structure, co-evolution, pharmacophore and torsional angles. Furthermore, DockNet is comparable to current state-of-the-art methods and, more importantly, demonstrates a novel deep learning architecture that breaks through limitations imposed by previous methods. Specifically, our method can generate predictions at much greater speeds than previous methods and is an effective method for high throughput studies at a proteome scale. Additionally, DockNet is trained to work with homology models as well as X-ray Crystal structures and can be applied in situations where accurate unbound protein structures cannot be obtained.
searchRunThis website is free for all users. For  policy please click here.
policy please click here.