1.Submission Page - Affinity prediction Mode
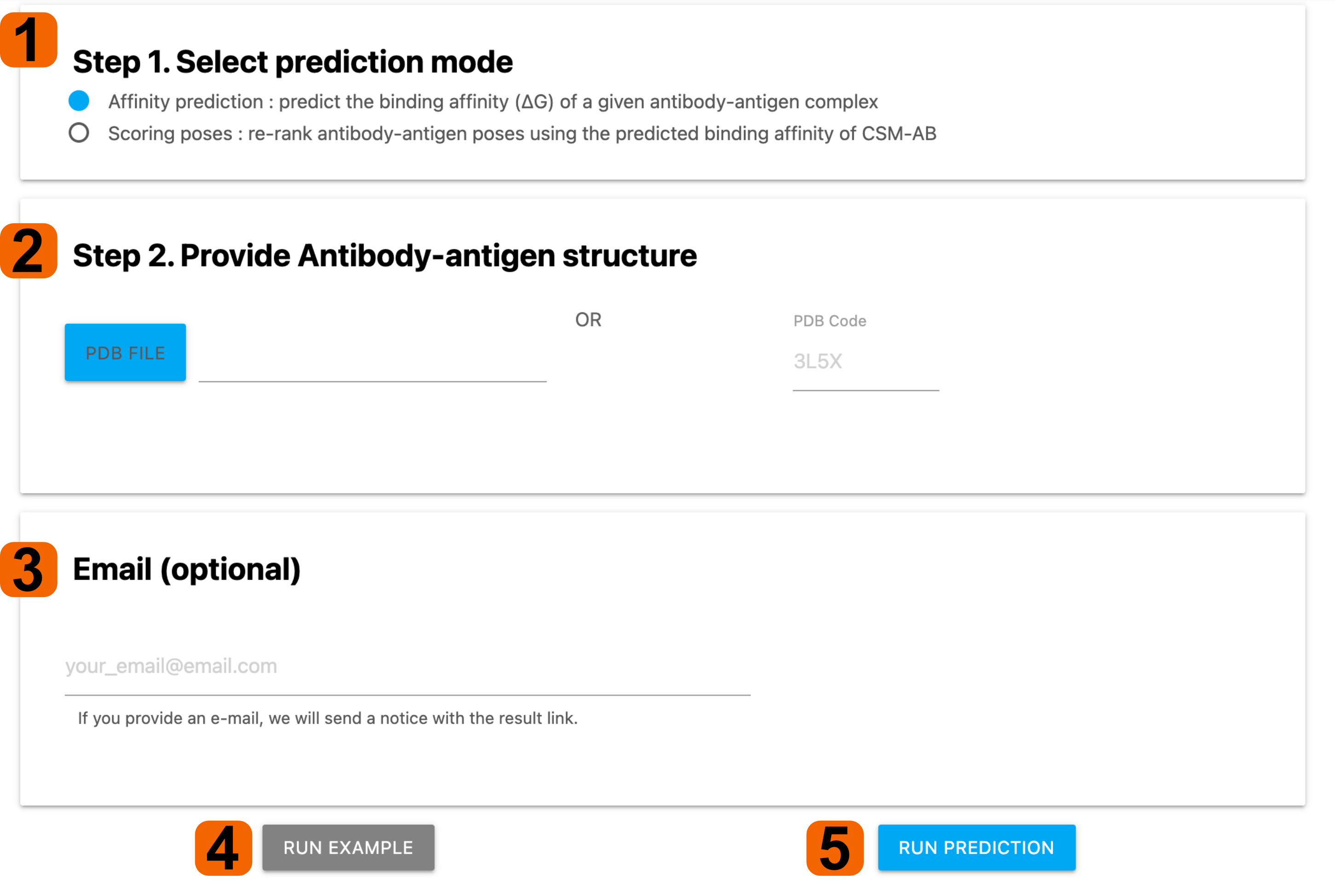
1. Prediction mode among Affinity prediction/Scoring poses needs to be
selected based on the purpose of the prediction.
- For Affinity prediction mode, users are required to provide an
Antibody-antigen complex structure for predicting the binding affinity.
2. An antibody-antigen complex can be provided by either of PDB file or PDB
code.
3. By providing an e-mail address, user can a notification email, after job
completion.
4. To visit example pages for each mode, users have to select the Affinity prediction mode
from step 1 and click 'Run Example' button.
5. Click RUN PREDICTION to get your prediction result.
2.Submission Page- Scoring poses Mode

1. Useres can score pre-docked poses via Scoring poses mode.
2. The Scoring poses mode accepts an Antibody (receptor) PDB and an Antigen (poses) PDB
which consits of upto 100 poses.
3. By providing an e-mail address, user can a notification email, after job
completion.
4. To visit example pages for each mode, users have to select the Affinity prediction mode
from step 1 and click 'Run Example' button.
5. Click RUN PREDICTION to get your prediction result.
3.Result Page- Affinity prediction Mode

The result page of Affiinity prediction Mode is divided into two sections:
1. The Results Table provides predicted bindin affinity (∆G), general information,
and the number of atomic interactions in bar plot.
2. Users can check the corresponding atomic interactions via
3D Visualisation
3. The 3D Visualisation provides atomic interactions of a given
Antibody-antigen complex. Using control panel, users are able to change the
representations of antibody and antigen separately. Each interaction can be turned
on/off using switches of which the colours are matched with the dashed-line in 3D viewer.
4. All information (interactions and predicted binding affinity) can be downloaded as
Pymol-session(PSE) and CSV files respectively.
4.Result Page- Scoring poses Mode
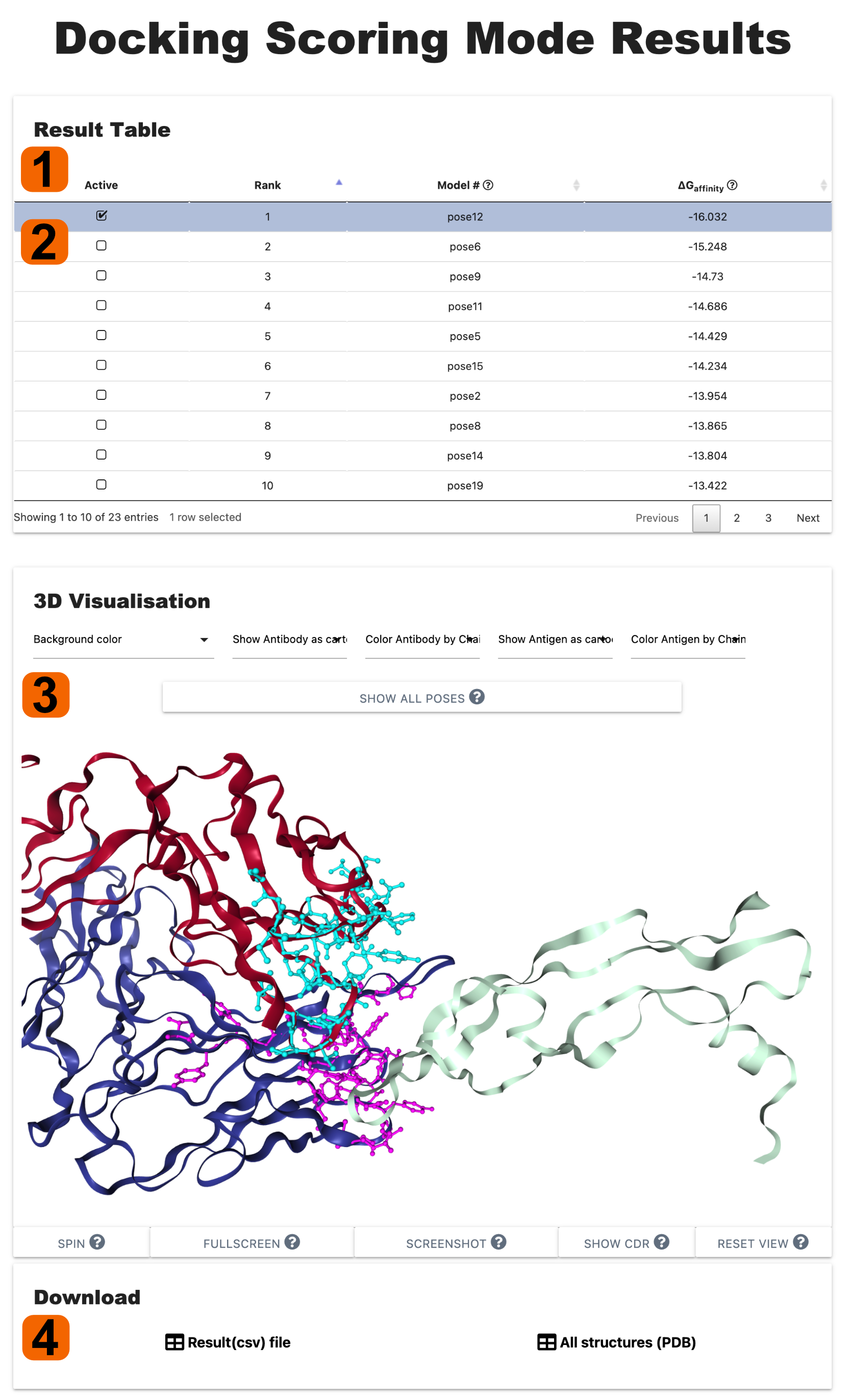
The result page of Scoring poses Mode is divided into two sections:
1. The Results Table provides predicted bindin affinity (∆G), pose number ,
and the Rank of poses.
2. Users can pick each complex and show in 3D Visualisation
3. The 3D Visualisation can display all poses or selected single complex from the result table.
4. All information (3D structures and result table) can be downloaded as
PDB and CSV files respectively.