VIVID: a web application for variant interpretation and visualisation in multidimensional analyses
Swapnil Tichkule,
Yoochan Myung,
Myo T. Naung,
Brendan R. E. Ansell,
Andrew J. Guy,
Namrata Srivastava,
Somya Mehra,
Simone M. Caccio,
Ivo Mueller,
Alyssa E. Barry,
Cock van Oosterhout,
Bernard Pope,
David B. Ascher,
Aaron R. Jex
Large-scale comparative genomics- and population genetics-based studies generate
enormous amounts of polymorphism data in the form of DNA variants. The ultimate goal of these studies
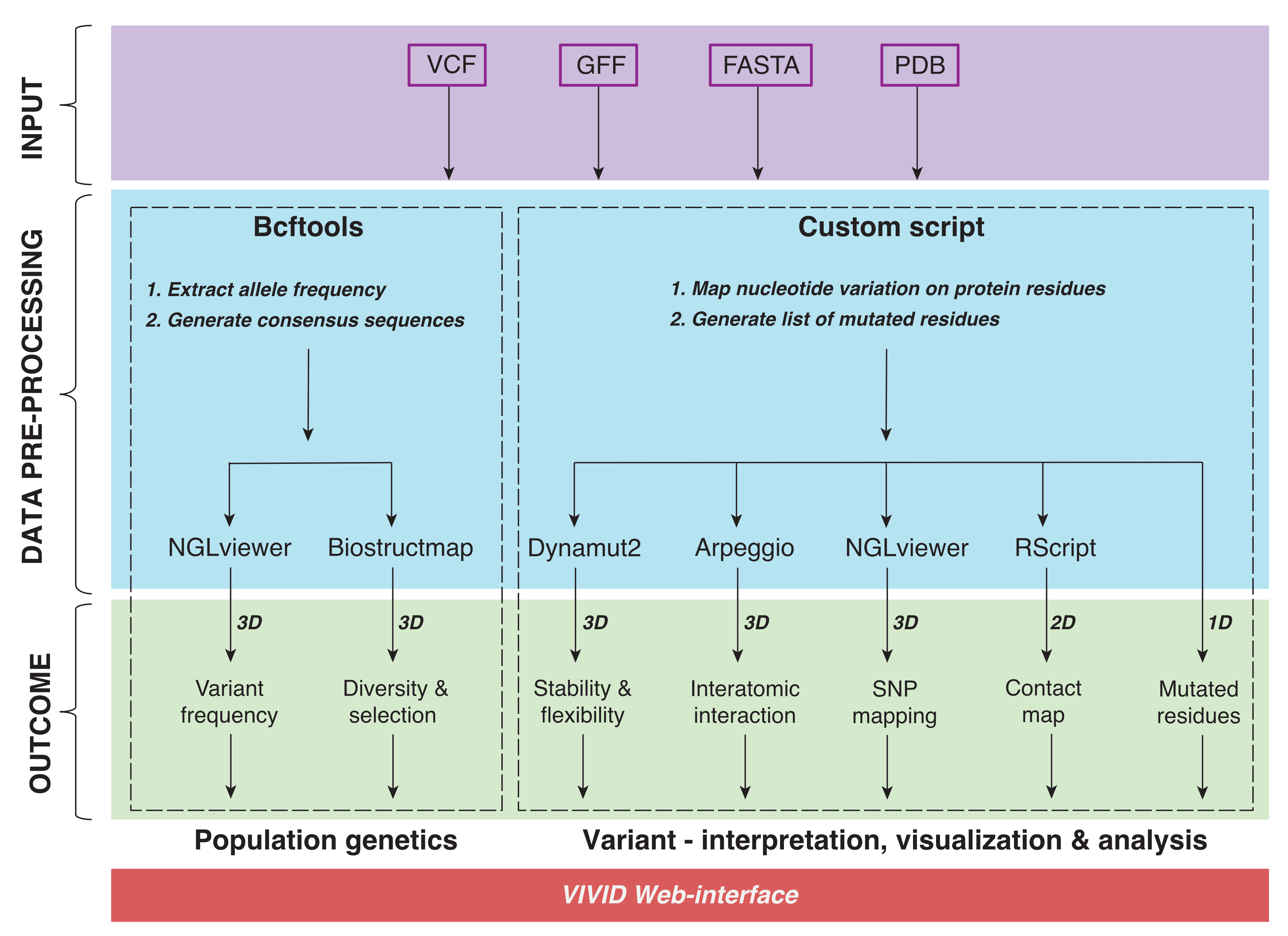
is to associate genetic variants with phenotypic outcomes. We introduce VIVID, an interactive,
user-friendly web application that integrates a wide range of approaches for encoding genotypic to
phenotypic information in any organism or disease, from an individual or population in three-dimensional (3D) space.
It allows mutation mapping and annotation, calculation of interactions and conservation scores,
prediction of harmful effects, population-scale genetic analyses and 3-dimensional (3D) visualisation
of genotypic information encoded in Variant Call Format (VCF) on AlphaFold2 protein models.
VIVID indeed offers a convenient way to rapidly assess genes of interest and accelerate research by
prioritising targets for experimental validation.