1. Predict: Submission Page
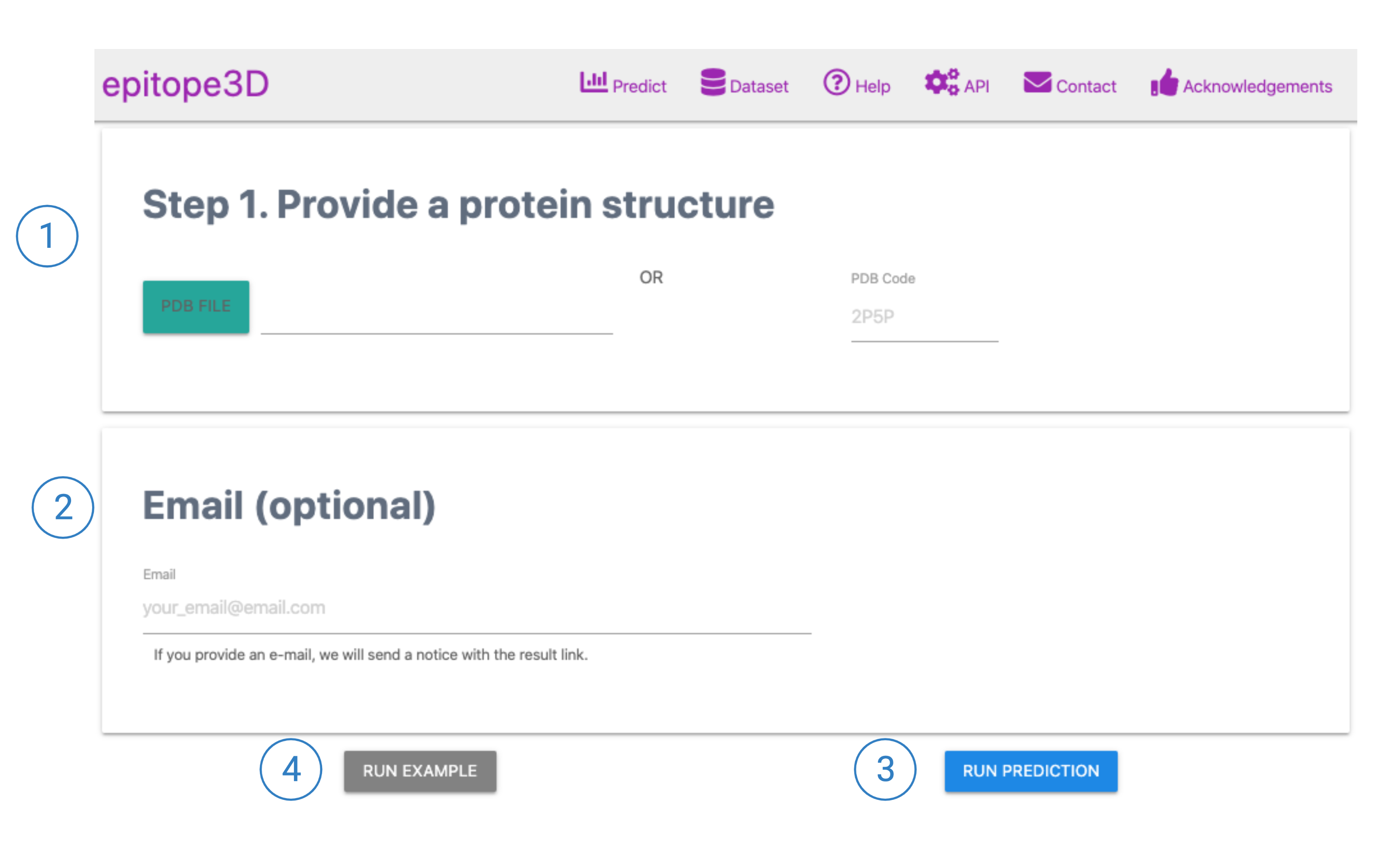
1. An Antigen structure can be provided by either of PDB file or PDB code.
2. Provide user's email (optional) if one would like to be advised when the prediction is finished.
3. Click RUN PREDICTION to submit the provided structure for prediction.
4. Instead, if user would like to view an example of Result's page, click 'Run Example' button.
2. Result Page: Conformational Epitope Prediction
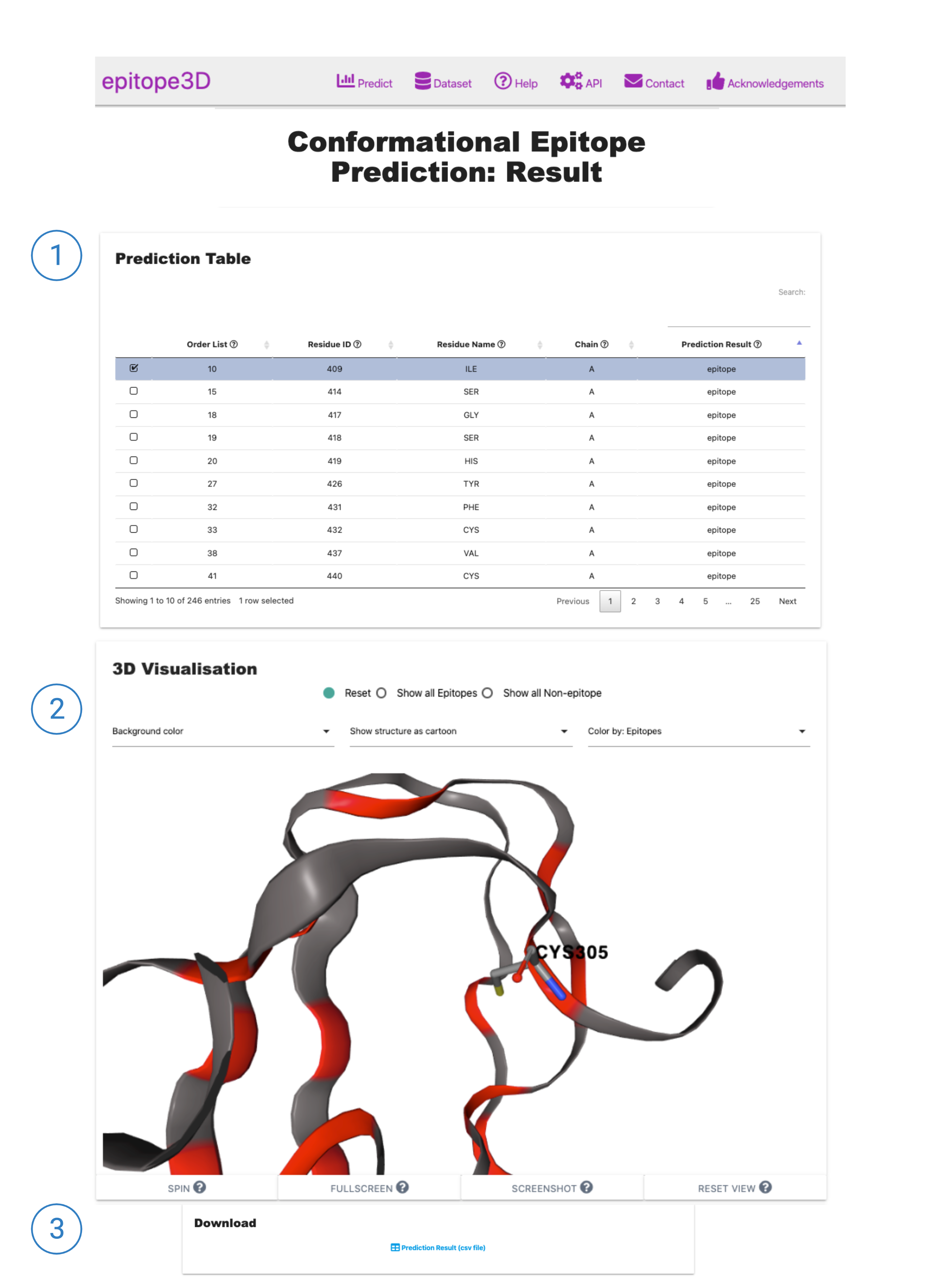
The result page is divided into three sections:
1. The Prediction Table section shows the output epitope prediction for each residue of the PDB file. The Order List column is a numerical list of residues in the same sequence as observed in the PDB file, followed by the Residue information (which is composed by the residue number (Residue ID), Residue Name and the Chain). The Prediction Result column stands for the classification outcome: epitope (means that the model predicted that residue as part of a conformational epitope), or non-epitope.
2. The 3D Visualisation provides a dynamic view of the PDB structure. Using control panel, users are able to change the representation of the antigen structure, for instance by coloring the epitope residues to distinguish them, select all the epitope residues at once, among other options.
3. The Prediction table is available to be downloaded as a .csv file.