HMMStruct: On use of tertiary structure characters in hidden markov models for protein fold prediction.
Ashar Malik, Caroline Puente-Lelievre, Nick Matzke, and David B. AscherAbstract: Innovations like AlphaFold and ESMFold have now theoretically made possible structure prediction of millions of proteins, most of which are arcane and poorly studied. The next logical step, therefore, is to predict the function of these novel protein structures. SCOP, CATH and ECOD are databases which group protein structure into families based on structure-function similarity. While predicted protein structures can be compared to already characterised structures, in these databases, these structures struggle to find suitable matches. An alternative thus is to use machine learning approaches for prediction, however these too struggle as within and across family structure comparisons can produce confounding results. In this work, we use tertiary characters generated by Foldseek to develop structure-based hidden markov models which are then used to classify novel protein structures. Results generate top-10 characterised matches for SCOP, CATH and ECOD using the conventional nearest neighbour search and the same number of results are returned searching against the models for each of the three databases with added GO functional annotations. Altogether, this work presents an important advance for rapid characterization of novel arcane structures.
Predict the fold of a structure
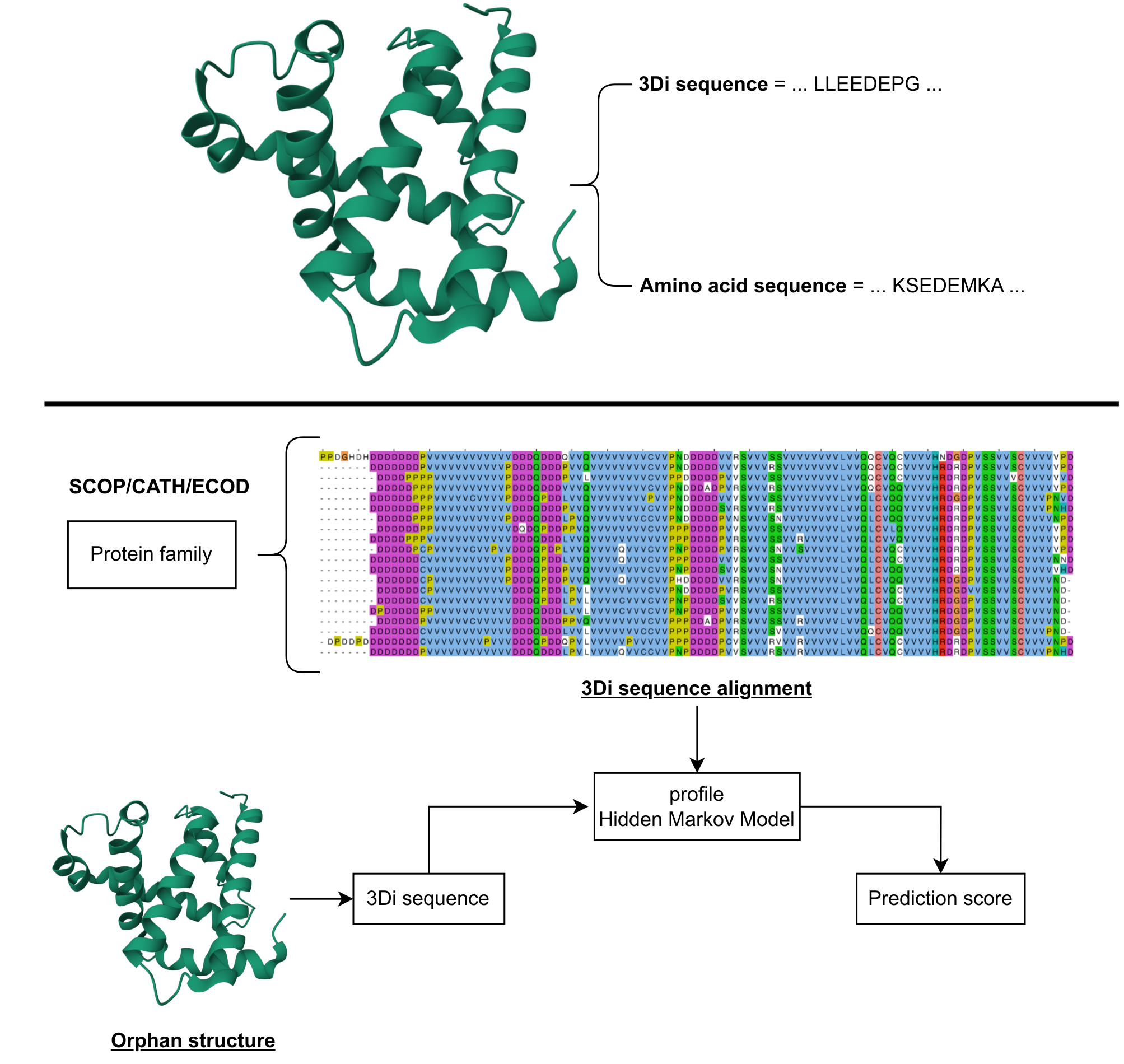
Protein structures are coverted to 3Di sequences using Foldseek. This resource compares the 3Di sequence of the query structures to SCOP/CATH/ECOD family specific profile HMMs and top-10 best scores are listed.